Figure 1.
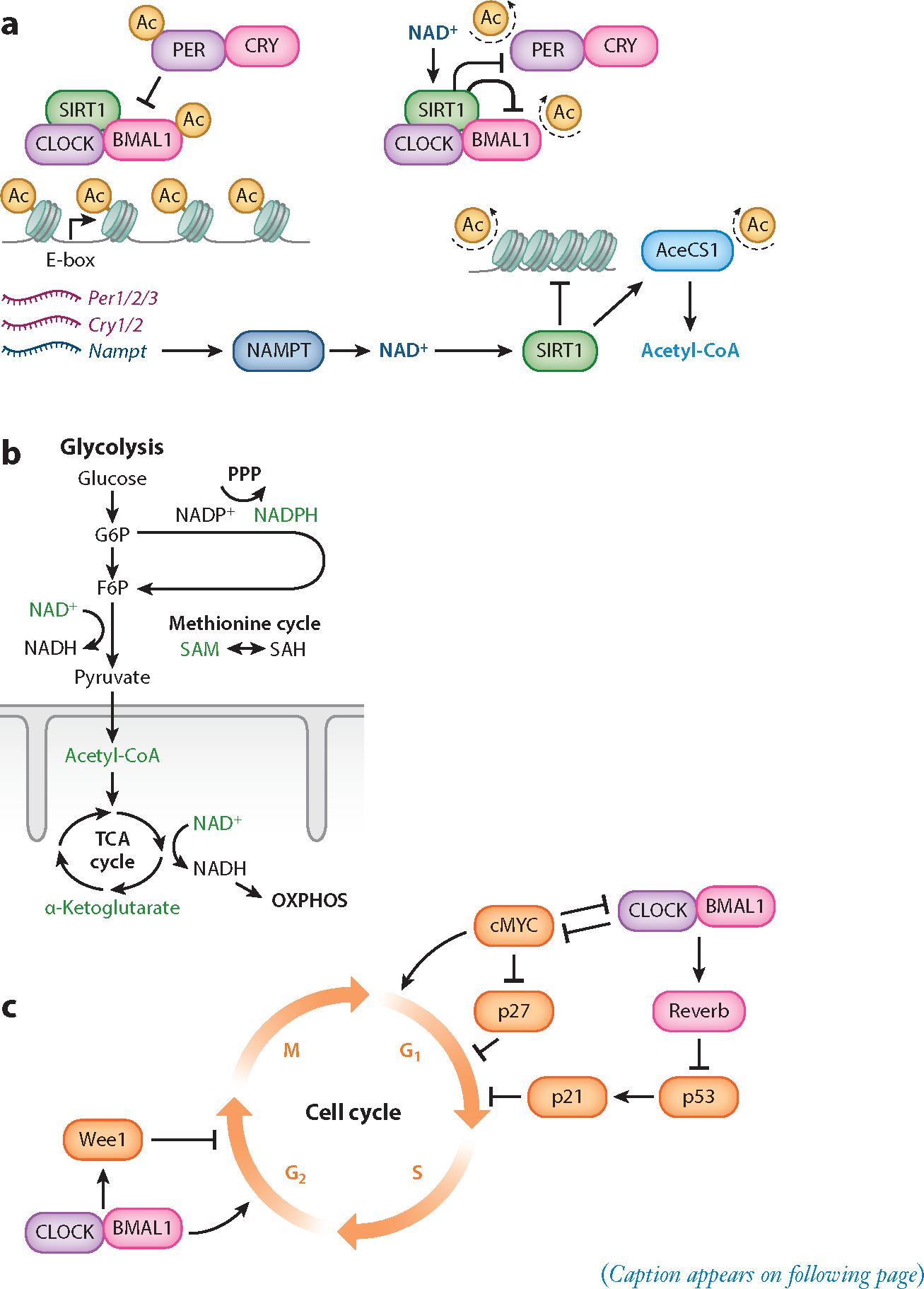
Circadian clock interaction with metabolism, cell cycle, and chromatin remodeling. (a) The core circadian TTFL is composed of the heterodimer CLOCK:BMAL1, which drives the transcription of their repressors PER1/2/3 and CRY1/2; these repressors dimerize in the cytoplasm and translocate to the nucleus to repress CLOCK:BMAL1. CLOCK:BMAL1 also drives the rhythmic accumulation of NAD+ by driving the transcription of the gene encoding the protein NAMPT, the rate-limiting enzyme in the NAD+-salvage pathway. The cyclic synthesis of NAD+ corresponds to the rhythmic activity of NAD+-dependent HDACs including SIRT1. Rhythms in SIRT1 activity over 24-hour periods also contribute to the cyclic synthesis of the universal acetyl donor acetyl-CoA through the enzyme AceCS1, which is activated by deacetylation. (b) Major catabolic pathways conserved across living cells include glycolysis, the TCA cycle, and OXPHOS. Each of these pathways produces metabolites that interact with circadian rhythms through the production of metabolites that act as substrates for posttranslational modifications or can activate protein-modifying enzymes (labeled in green). Among anabolic pathways, the PPP and the methionine cycle generate metabolites that interact with circadian rhythms. (c) Prominent connections of the circadian clock (purple) with the cell cycle (orange), emphasizing the checkpoints that are especially important for their coupling in stem cells. Abbreviations: Ac, acetyl; AceCS1, acetyl-CoA synthetase 1; CRY, Cryptochrome; F6P, fructose 6-phosphate; G6P, glucose 6-phosphate; HDAC, histone deacetylase; NAD+, nicotinamide adenine dinucleotide; NADP+, nicotinamide adenine dinucleotide phosphate; NAMPT, nicotinamide phosphoribosyltransferase; OXPHOS, oxidative phosphorylation; PER, Period; PPP, pentose phosphate pathway; SAH, S-adenosylhomocysteine; SAM, S-adenosyl methionine; TCA, tricarboxylic acid; TTFL, transcriptional-translational feedback loop.
