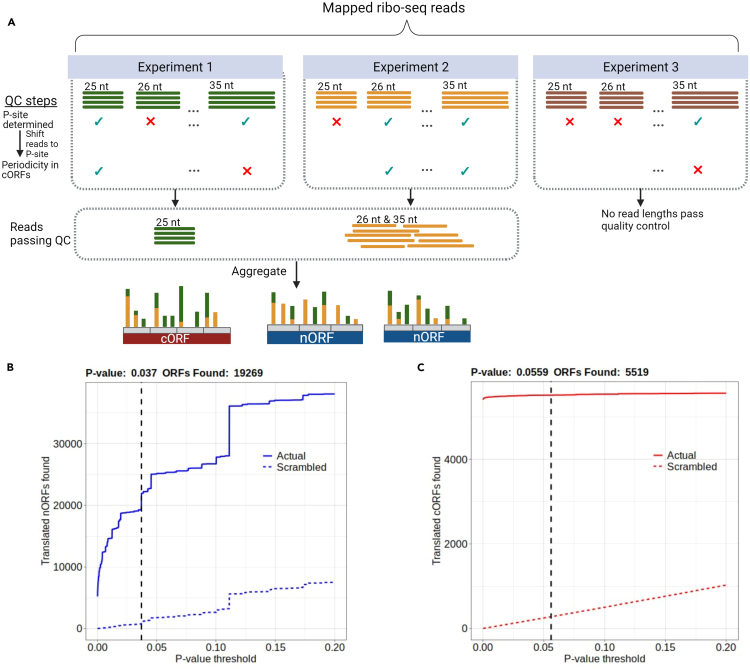
Figure 1.
Thousands of noncanonical S. cerevisiae ORFs identified by iRibo
(A) The process by which iRibo generates a translation profile using ribo-seq data. Mapped ribo-seq reads from each experiment are grouped by read length, and for each length the P-site is inferred. Reads are then shifted to the P-site. For quality control, three-nucleotide periodicity is then checked among canonical ORFs. For each read length in which a P-site can be inferred, and which shows periodicity among canonical ORFs, all reads are then aggregated in the genome to allow determination of which ORFs are translated.
(B) Translated nORFs found by iRibo at a range of p-value thresholds. A 5% false discovery rate was identified at a p-value threshold of 0.037 for nORFs, indicating 19269 translated nORFs. The dashed blue line represents the average number of nORFs discovered performing the same test on 100 scrambled distributions of reads.
(C) Translated cORFs found by iRibo at a range of p-value thresholds. At the p-value threshold of .037, set to obtain a 5% FDR among nORFs, 5519 cORFs are detected. The dashed red line represents the average number of cORFs discovered performing the same test on 100 scrambled distributions of reads.