Figure.
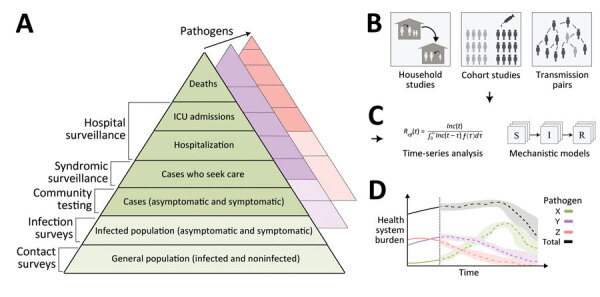
An integrated surveillance system for multiple circulating respiratory pathogens. A) Surveillance systems, which collect data continuously, monitor changes in infection outcomes (or behavior) at different levels of severity on the severity pyramid. In general, the proportions in each level can vary over time (e.g., because of emerging new variants), so surveillance should be undertaken at multiple levels. Collecting additional behavioral data can help disentangle changes in the data streams resulting from behavior specifically (e.g., changes in healthcare-seeking behavior) or indirectly (e.g., changes in transmission rates due to changes in contact rates). In addition, collecting data on interventions (e.g., vaccine uptake, therapeutic use) at all levels of the severity pyramid can help to enable estimates of intervention effectiveness. In general, multiple pathogens will circulate concurrently, and it may be necessary for surveillance systems to be extended to distinguish between the different pathogens. Integrating genomic sequencing of samples collected at all levels can additionally allow differences in dynamics between variants to be identified. B) Enhanced surveillance activities conducted periodically can augment continuous surveillance. Enhanced surveillance activities may inform on key epidemiologic quantities, such as the generation interval and vaccine effectiveness, which are important for predicting future transmission dynamics. C) Data collected from different sources of clinical, epidemiologic, and genomic surveillance can be synthesized through data analysis pipelines and used to generate epidemic forecasts and scenario projections (long- and short-term). D) Forecasts and projections can be used to support public health decision-making and planning. For example, they can be used to predict the timing and size of concurrent epidemics of influenza, SARS-CoV-2, and RSV and anticipate the resulting combined healthcare burden. ICU, intensive care unit; S→I→R, susceptible-intermediate-resistant model.
