Fig. 5: Inactivation of SREBPs inhibits lipogenesis in HTM cells.
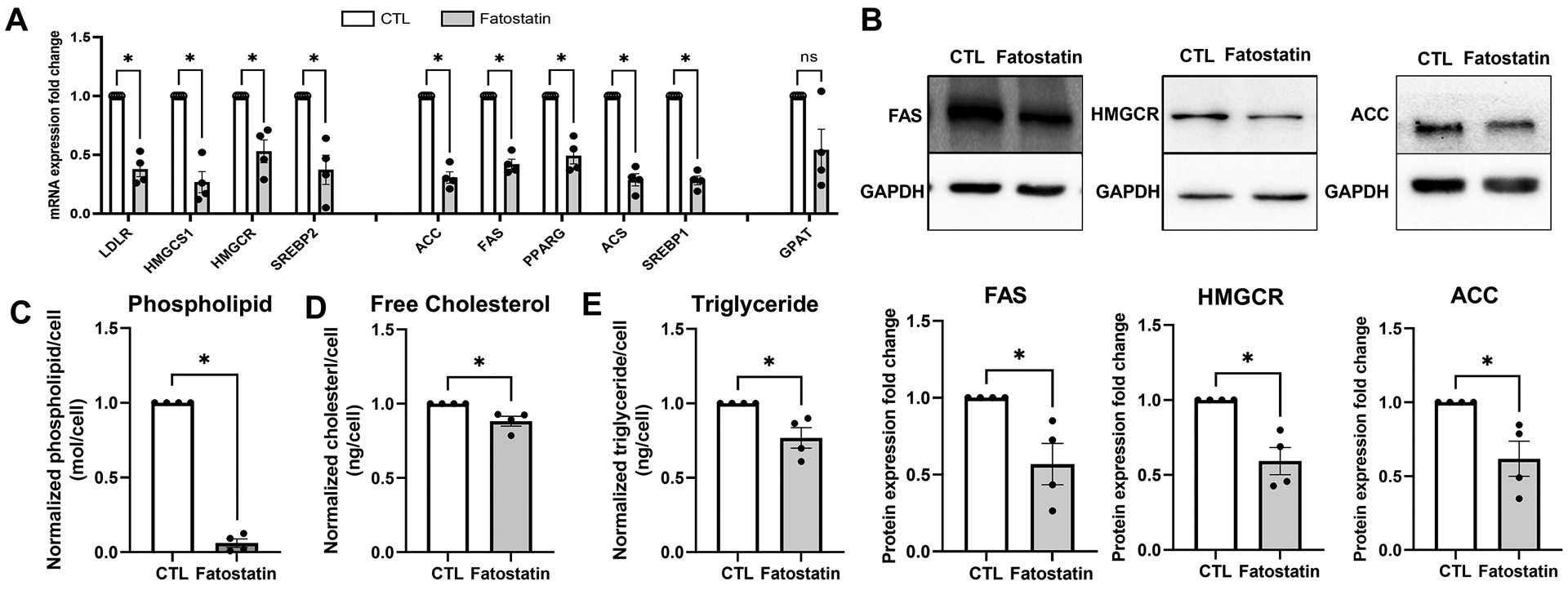
(A) Fatostatin treatment significantly decreased SREBPs responsive genes mRNA expression involved in 1) cholesterol biosynthesis: low-density lipoprotein receptor (LDLR), hydroxymethylglutaryl-CoA synthase 1 (HMGCS1), 3-hydroxy-3-methylglutaryl-CoA Reductase (HMGCR), SREBP2; 2) fatty acid biosynthesis: acetyl-CoA carboxylase (ACC), fatty acid synthase (FAS), SREBP1, peroxisome proliferator- activated receptor gamma (PPARG), acetyl-CoA synthetase (ACS); 3) triglyceride and phospholipid biosynthesis: glycerol-3-phosphate acyltransferase (GPAT). 18S were used as internal controls for qPCR analysis. (B) Immunoblotting shows that fatostatin significantly reduced FAS, HMGCR, and ACC protein expression compared to the control. GAPDH was used as loading control. (C), (D) and (E) show that fatostatin significantly reduced total lipid biosynthesis in HTM cells, including normalized phospholipid/cell, normalized cholesterol/cell, and normalized triglyceride/cell. Values represent the mean ± SEM, where n = 4 (biological replicates). *p < 0.05 was considered statistically significant.
