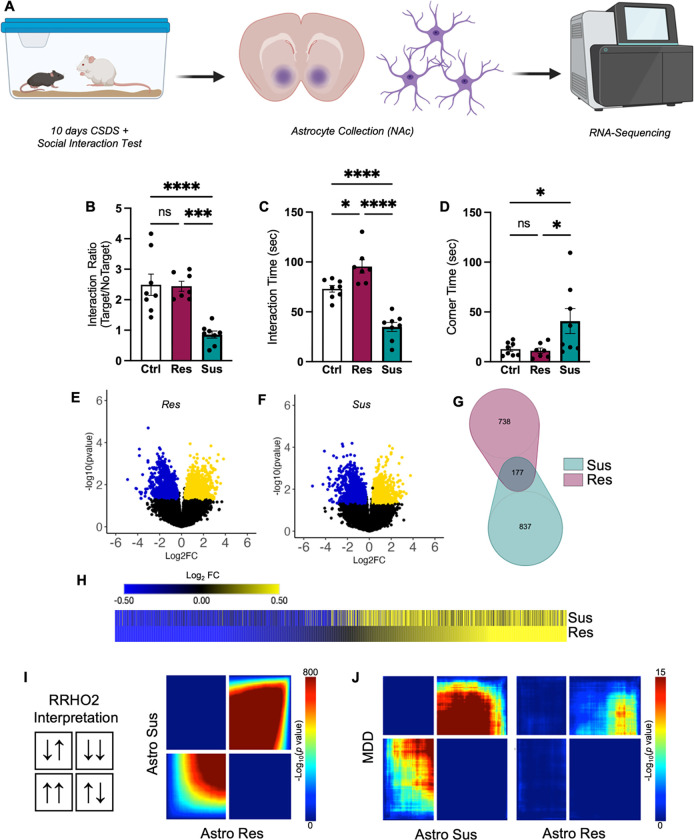
Figure 1. Astrocyte transcriptome in NAc responds robustly to chronic stress.
A) Cartoon illustration of experimental design. B–C) Behavior of phenotyped animals selected for astrocyte collection and RNA-seq. CSDS resulted in Res (purple) and Sus (teal) phenotypes, as expected. B) Sus animals demonstrate decreased Interaction Ratios compared to Ctrl and Res animals (F(2,20) = 15.63, P < 0.0001; Tukey post-hoc ***p < 0.001). C) Res animals demonstrate an increase in time interacting with a novel CD1 aggressor compared to Ctrl and Sus animals, while Sus animals demonstrate a decrease compared to Ctrl animals (F(2,20) = 38.44, P < 0.001; Tukey post-hoc *p < 0.05; ****p < 0.0001). D) In contrast, Sus animals demonstrate an increase in time spent in corners compared to both Res and Sus (F(2,20) = 4.636, P = 0.0222; Tukey post-hoc *p < 0.05). Volcano plots of detected genes from E) Res and F) Sus astrocytes compared to Ctrl. Upregulated DEGs are indicated in yellow, while downregulated DEGs are indicated in blue. G) Venn diagram reveals little overlap between significant DEGs in Res (purple) and Sus (teal) astrocytes. However, H) union heatmap demonstrates considerably similar Log2FC expression of significant DEGs where again upregulated DEGs are indicated in yellow and downregulated DEGs are indicated in blue. The union heatmap nevertheless does highlight DEGs that are regulated differently in Res versus Sus. I) RRHO2 threshold-free genome-wide comparison confirms the union heatmap by revealing partly concordant gene expression between Res and Sus. J) RRHO2 comparison of human MDD bulk RNA-seq of NAc to Sus (left) and Res (right) RNA-seq of NAc astrocytes demonstrates some concordant expression between the mouse and human datasets. Data represented at mean +/− SEM, n = 7–8 animals per condition. RNA-seq: RNA-sequencing; CSDS: chronic social defeat stress; Res: resilient; Sus: susceptible; Ctrl: control; DEG: differentially expressed genes; Log2FC: log2 fold change; RRHO2: rank rank hypergeometric overlap.