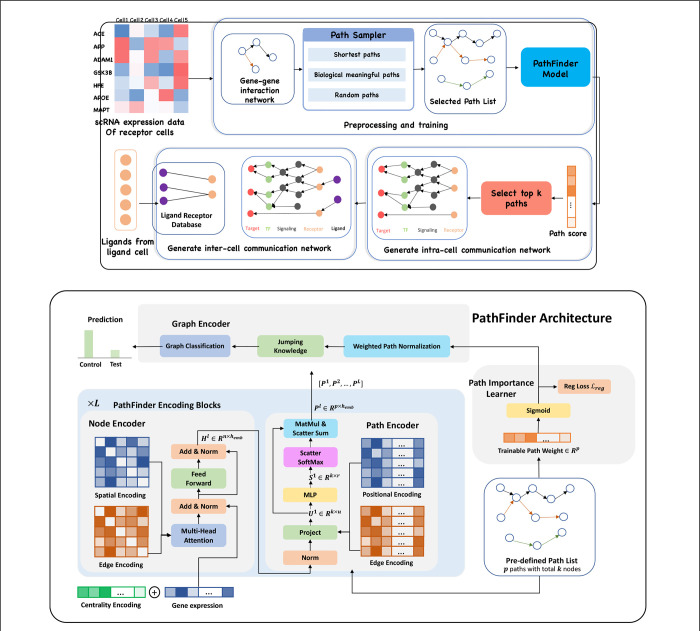
Figure 1. (Upper) Overview of PathFinder method to discover both intra- and inter-cell communication network.
The input scRNA expression data with both samples from the control condition and the test condition is used to construct the gene-gene interaction network based on our large database. Next, the path sampler is used to generate all pre-defined path from the interaction network. Then, the PathFinder model is trained to separate the cells from two different conditions. After the training, the learned path score can indicate the importance of each path. Next, the top paths are selected to generate the intra-cell communication network. Finally, the Ligand-receptor database is used to link all picked ligands (like differential expressed ligands) from ligand cells to the receptors in the intra-cell communication network of receptor cells to construct the inter-cell communication network. (Lower) Model architecture of PathFinder. The PathFinder consists of three components: node encoder, path encoder, and graph encoder. The node encoder is a stack of layer of transformer with special encoding to encode local graph structure information of each node. The path encoder take the output from each layer of node encoder to learn long-range path embedding for each pre-defined path. Finally, the graph encoder aggregate information from each path to generate graph embedding and make final prediction. In the graph encoder, the trainable path weight will be learned to assign each path an importance score, which can be used to generate intra-cell communication networks.