Figure 1. Obelisk alpha has a predicted extensive secondary structure and appears to colonise and speciate within the human gut.
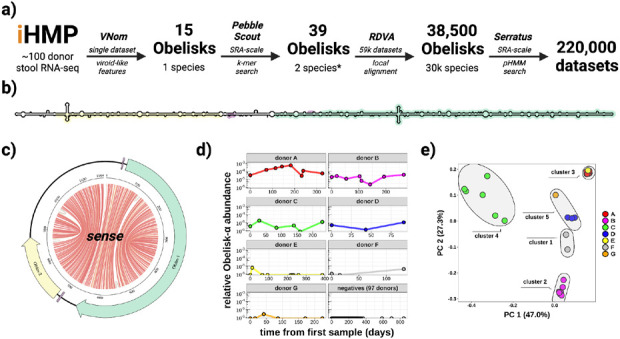
a) overview of the iterative approach taken in Obelisk discovery, (see methods) b) schematic of the predicted sense consensus secondary structure derived from all non-redundant, 1164 nt Obelisk-αs found using SRA-scale k-mer matching (PebbleScout). Predicted open reading frames (ORFs) 1 and 2 (green/yellow), and Shine-Delgarno sequences (purple) shown, c) “jupiter” plot of Obelisk-α coloured as in b), chords illustrate predicted basepairs (basepair probabilities grey, 0.1, to red, 1.0) d) Obelisk-α relative read abundance for six donors (A-G); sequence data from in Lloyd-Price et al., 2019 and time in days from first sample. e) Principal component analysis of sequence variation seen in Obelisk-α reads in Lloyd-Price et al., 2019 (the initial iHMP dataset), grouped by k-means clustering with 5 centres, coloured as in d).
