Figure 2. Obelisks encode putatively well-folded proteins.
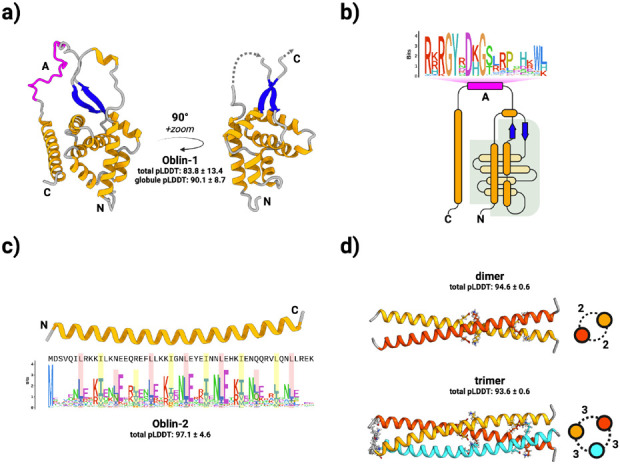
a) Obelisk open reading frame 1 (Oblin-1) is predicted (total mean-pLDDT ± SD = 83.8 ± 13.4, see methods) to fold into a stereotyped N-terminal “globule” formed of a three alpha helix (orange) bundle partially wrapping around an orthogonal four helix bundle, capped with a beta sheet “clasp” (blue, globule mean-pLDDT = 90.1 ± 8.7), joined by an intervening region harbouring the conserved domain-A (magenta) with no predicted tertiary structure, to an arbitrarily placed C-terminal alpha helix. “Globule” emphasised on the right. b) a to-scale (secondary structure) topological representation of Oblin-1 with the “globule” shaded in grey, and the domain-A emphasised with this bit-score sequence logo (see methods). c) Obelisk Oblin-2 is confidently predicted (mean-pLDDT = 97.1 ± 4.6 ) to fold into an alpha helix which appears to be a leucine zipper. Sequence logo of an “i+7” leucine spacing emphasised in red, with hydrophobic “d” position residues emphasised in yellow (expanded in Supplementary Figure 4b). d) homo-multimer predictions of Obelisk-alpha Oblin-2. top: dimer (mean-pLDDT = 94.6 ± 0.6), bottom: trimer (mean-pLDDT = 93.6 ± 0.6). Side-on representations of homomultimers shown with numbers of inter-helix salt-bridges (see Supplementary Figure 5).
