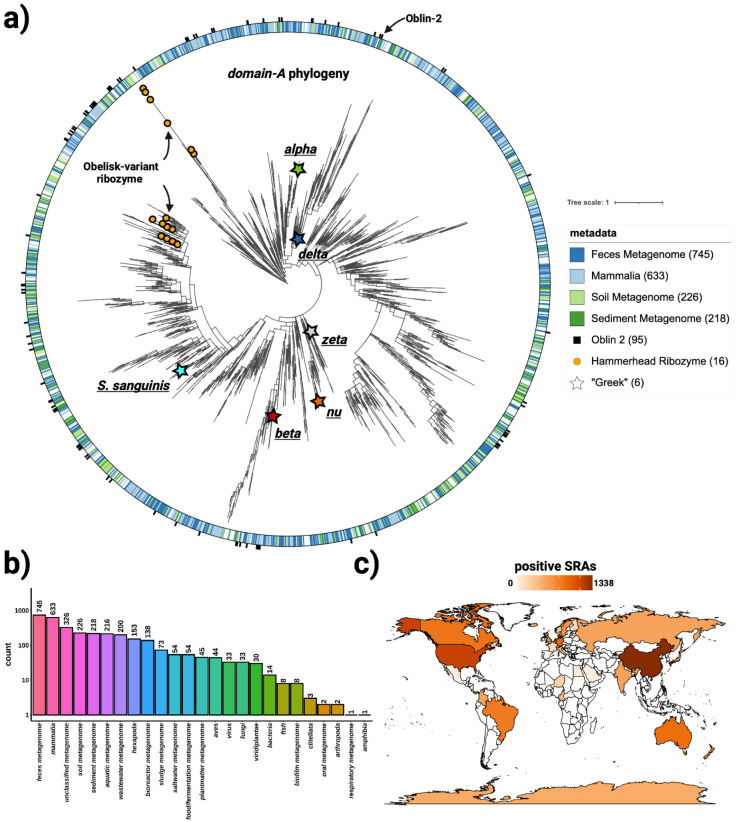
Figure 3. Obelisks form their own globally distributed phylogenetic group.
a) a maximum likelihood, midpoint-rooted, phylogenetic tree (see methods) constructed from a non-redundant set of 3265 Serratus and RDVA domain-A sequences, with RDVA genomes positive for Obelisk-variant self cleaving Hammerhead Type III ribozymes illustrated as orange circles on leaves, and the top four known classes of SRA “host” metadata depicted as the colour band (see legend), and with per-RDVA-genome co-occurrence of Oblin-2 (based on blastp hits against the Oblin-2 consensus) illustrated as the outer ring (black studs). Leaves that correspond to domain-A sequences from Figure 4 are illustrated with stars. b) Counts of non de-replicated SRA datasets used to construct a) sorted by their “host” metadata; we note that “host” metadata likely fails to account other organisms’ genetic material that was sequences alongside the “host” (e.g. signals from these hosts’ microbiomes maybe be detected in tandem). c) Counts of non de-replicated SRA datasets used to construct a) arranged by sample geolocation (where known) illustrated on a world map (darker orange = more SRA datasets contributed to a)). We note that SRA counts are not expected to correlate with true geo-/ecological prevalence, but are still indicative of global presence.