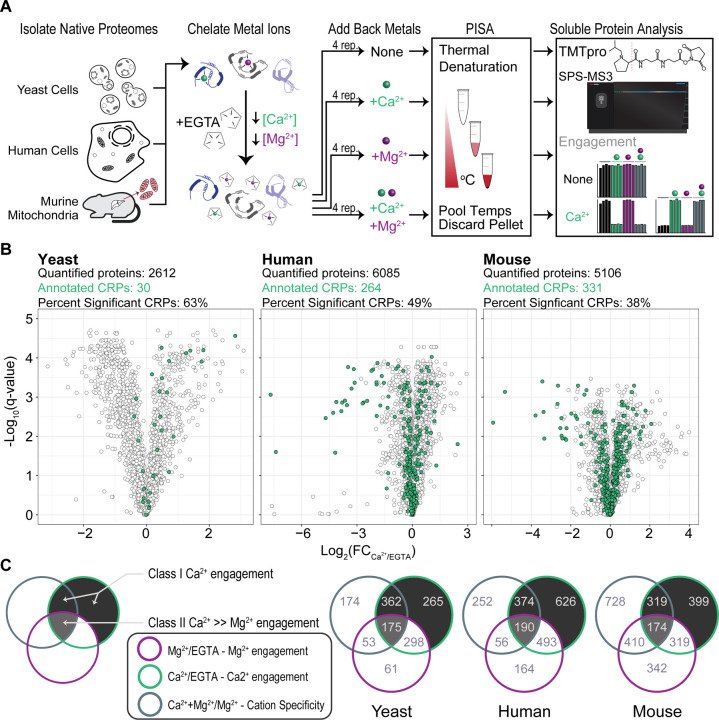
Figure 1:
PISA with isobaric sample multiplexing in three different proteomes identifies Ca2+-regulated proteins.
(A) Schematics of the experimental workflow to detect ion-induced changes in protein thermal stability. Lysates from yeast, human and isolated murine liver mitochondria were treated with chelator EGTA. After the addition of ions, a temperature gradient was applied to aliquoted samples, the samples were then mixed, and insoluble (heat denatured) material was precipitated. Soluble proteins were labeled with isobaric tags, and protein abundance was quantified.
(B) Volcano plots show protein abundance in Ca2+ relative to EGTA, and their q-values. Proteins with a |log2(FC)| ≥ 0.2, and q-value ≤ 0.05 (Welch’s t-test with multiple hypothesis correction) were considered to show significant thermal stability changes. Green dots indicate proteins with previous Ca2+ annotation.
(C) Venn diagrams show proteins with significant thermal stability changes in each category listed. Proteins that are in the shaded compartments represent CRPs. Proteins that do not show a significant thermal stability change between Mg2+ and Mg2+Ca2+ conditions were considered to be non-specific ion responders and excluded from the list of CRPs.