Abstract
In polyomaviruses the pentameric capsomers are interlinked by the long C-terminal arm of the structural protein VP1. The T=7 icosahedral structure of these viruses is possible due to an intriguing adaptability of this linker arm to the different local environments in the capsid. To explore the assembly process, we have compared the structure of two virus-like particles (VLPs) formed, as we found, in a calcium-dependent manner by the VP1 protein of human polyomavirus BK. The structures were determined using electron cryomicroscopy (cryo-EM), and the three-dimensional reconstructions were interpreted by atomic modeling. In the small VP1 particle, 26.4 nm in diameter, the pentameric capsomers form an icosahedral T=1 surface lattice with meeting densities at the threefold axes that interlinked three capsomers. In the larger particle, 50.6 nm in diameter, the capsomers form a T=7 icosahedral shell with three unique contacts. A folding model of the BKV VP1 protein was obtained by alignment with the VP1 protein of simian virus 40 (SV40). The model fitted well into the cryo-EM density of the T=7 particle. However, residues 297 to 362 of the C-terminal arm had to be remodeled to accommodate the higher curvature of the T=1 particle. The loops, before and after the C-terminal short helix, were shown to provide the hinges that allowed curvature variation in the particle shell. The meeting densities seen at the threefold axes in the T=1 particle were consistent with the triple-helix interlinking contact at the local threefold axes in the T=7 structure.
The BK virus (BKV) is a human virus belonging to the Polyomaviridae family. It is a nonenveloped virus (∼50.0 nm in diameter) with a circular double-stranded DNA genome (∼5 kb). The capsid has icosahedral symmetry and is built of 72 capsomers that are all pentamers of the protein VP1 arranged in a T=7 icosahedral lattice (21). All known polyomaviruses have three structural proteins (VP1, VP2, and VP3), of which VP1 is the major capsid protein. Overall amino acid sequence homology between BKV and the other human polyomavirus, JCV, is 75%, and that with the simian polyomavirus (SV40) is 69% (9). In the VP1 protein the sequence similarity rises to 77% and 74% for the JCV and SV40, respectively (35). Due to the high similarity, the solved atomic structure of VP1 of SV40 provides us a template to create a model of the BKV VP1 protein folding.
The structures of the SV40 and murine polyomavirus have been determined and show similar features to that seen in the BKV (1, 12). The VP1 pentamer of SV40 and murine polyomavirus is built as a ring of five β-barrel-shaped VP1 monomers, tightly linked by interacting loops between the framework of β-strands (22, 33, 34, 40). The C-terminal subdomain of each VP1 monomer “invades” a neighboring pentamer, thereby tying the pentamers together in the virion shell. There are six unique monomers building up the capsid (monomer α, α′, and α" at the local threefold; β, β′ around the icosahedral threefold, and γ at the twofold) (34). The major structural differences between the six unique monomers are found in their C termini and are essential for the formation of the icosahedral capsid. The most flexible region seemed to be the outermost C terminus of the peptide chain (amino acids 355 to 364). The structure of this region has not been defined in three of the six unique monomers (α, α", and β).
Under certain conditions the polyomavirus capsid disassembles into pentamers, which can be reassembled. The reassembled particle may have different configurations and sizes depending on the buffer conditions. This was shown for recombinant SV40 VLPs (17) and recombinant murine polyoma VLPs (28, 29). In the latter case, Salunke et al. (29) demonstrated that three differently sized VLPs appeared, depending on the buffer conditions. They concluded through computational modeling that two of the reassembled murine polyomavirus particles had icosahedral symmetry and were composed of 12 and 72 pentamers, in a T=1 and T=7, respectively, surface lattice. The third particle was composed of 24 pentamers with an octahedral symmetry.
Many studies have demonstrated that calcium ions play an important role in viral assembly (3, 5, 6, 13, 14, 17, 20, 25, 26, 28). Besides the calcium-binding site, disulfide bonds have also been found to be involved in maintaining capsid stability of the polyomavirus (5, 6, 11, 14, 15, 19, 30, 36). Interpentameric disulfide linkages have been revealed by X-ray crystallography in the capsid structure of SV40 and murine polyomavirus (22, 33, 34).
The gene of the major structural protein of the BKV VP1 was recently expressed in Tn5 cells using a recombinant baculovirus vector (21). The BKV VP1 self-assembled into VLPs in the nucleus, and the particles were then efficiently released into the culture medium. These VLPs possessed similar antigenicity as native BKV particles and were indistinguishable as negative-stained specimens in electron microscopy. The structure was determined to 2.0 nm resolution, using electron cryomicroscopy (cryo-EM) and three-dimensional reconstruction, and showed similar features to both SV40 and murine polyomavirus.
Here, we report for the first time the three-dimensional structure of a smaller BKV VLP determined by cryo-EM, image reconstruction, and docking of the VP1 model. We compare the structure of the two different BKV VLPs and explore the in vitro disassembly/reassembly process of them.
MATERIALS AND METHODS
Expression and purification of the BKV capsid protein VP1.
For the large-scale expression of the BKV capsid protein VP1 (21, 37), an insect cell line from Trichoplusia ni, BTL-Tn 5B1-4 (Tn5) (Invitrogen, San Diego, CA) was used. Tn5 cells were infected with recombinant baculoviruses at a multiplicity of infection (MOI) of 10 and were incubated in EX-CELL 405 medium (JRH Biosciences, Lenexa, KS) for 7 days at 26.5°C. Intact cells, cell debris, and progeny baculoviruses were removed by centrifugation at 10,000 × g for 90 min. The supernatant was then spun at 25,000 rpm for 2 h in a Beckman SW28 rotor. The resulting pellet was resuspended in 4.5 ml of EX-CELL 405 at 4°C overnight. After mixing with 2.1 g of CsCl, the sample was centrifuged at 35,000 rpm for 24 h at 4°C in a Beckman SW50.1 rotor. Four bands were harvested by puncturing the tubes with a 22-gauge needle. To remove the CsCl, each band was diluted 10× and centrifuged for 2 h in a Beckman TLA55 rotor at 50,000 rpm; the pellet was then resuspended in 150 mM NaCl, 10 mM TRIS-HCl, pH 7.4.
Dialysis experiments.
To screen for factors effecting dissociation and reassembly of the BKV VLPs, a homemade dialysis equipment that allowed dialyses of small amounts of sample (20 to 40 μl) was used. BKV VLPs (35 μl, 0.5 mg/ml) were first dialyzed against different concentrations of EDTA (10 to 100 mM), 2-mercaptoethanol (2-ME) (10 to 100 mM), and NaCl (0.15 to 1 M) in an attempt to dissociate the VLPs. When the VLPs had been dissociated, the sample (now containing free pentamers) was again dialyzed against buffers with different concentrations of a monovalent salt, NaCl (0.0 to 1.0 M), pH (7.0 to 7.8), and a divalent ion, Ca2+ (0.0 to 100.0 mM), to make the pentamers reassemble into VLPs. All experiments were done in room temperature and overnight. The selection of conditions for the second dialysis, like combinations of different pH, calcium, and NaCl concentrations, was done with the aid of the software DESIGN (32).
To determine the proportion of small (26.4 nm in diameter) and large VLPs (50.6 nm in diameter) in the different reassembly experiments, samples were applied to carbon-coated grids, negatively stained, and checked by EM. Thirty fields were randomly chosen from each grid and micrograph taken. In each the numbers of small and large VLPs were counted.
Negative contrasting electron microscopy.
For EM, a sample of ∼3 μl was applied to the charged grid and allowed to settle for 20 seconds. The solution was removed by blotting with a filter paper and the sample washed once with 3 μl of water for 15 seconds before stained with a 2% uranyl acetate solution. After 15 seconds the uranyl acetate solution was blotted off with a filter paper. The samples were observed by a Philips CM120 electron microscope.
Cryo-electron microscopy and three-dimensional image analysis.
Frozen-hydrated specimens were prepared by applying 3.0-μl droplets of an aqueous mixture of reassembled BKV VLPs (1.0 to 1.5 mg/ml) on 300-mesh copper grids coated with holey carbon film. The images of the frozen VLPs were recorded with a Philips CM120 (Philips Electronics Instruments) by defined low-dose condition. The micrographs were taken with Kodak SO163 films (Eastman Kodak Co., New York, NY) at 45,000 nominal magnifications and 120 kV operating voltage. Each area of specimen was recorded twice as focal pairs with defocus value of 1.0 and 3.0 μm, respectively (38). Micrographs with sufficiently separated and well-distributed particles, exhibiting minimal astigmatism, were digitized at 14-μm intervals (0.311-nm sampling at the specimen) with a Zeiss microdensitometer. Individual particle images were extracted and analyzed with icosahedral symmetry processing procedures to reconstruct the three-dimensional structure (39). Computations were performed with interactive FORTRAN programs on Alphastations (Digital Equipment Co., MA).
The initial phase origins of selected particle images were obtained by using a cross-correlation method, where the particle orientations were determined through modified self-common-lines and polar Fourier transform procedures (2, 10). This was followed by interparticle orientation refinement with increasing numbers of unique images by cross-common-lines procedures. To improve the sensitivity and reliability of the orientation refinement procedures with the BKV VLPs images, the data were Fourier-filtered to remove both low- and high-frequency noise beyond the processing regions. Refinement of origins and orientations was repeated in cycles at progressively higher spatial frequencies until no further improvement was found in the common-lines phase residues. Besides the procedures described above, back-projection images of the preliminary reconstruction were used as references to accurately refine the phase origins and orientations of the corresponding images. The resolution was progressively improved and the final three-dimensional reconstruction was computed to a resolution of 2.4 nm, which was within the limit imposed by the first zero of the contrast transfer function of the electron microscope.
Fitting of BKV VP1 model into the EM density map.
The atomic model of BK VP1 protein was constructed from SV40 VP1 using the SWISS-MODEL Protein Modeling Server (31) and the software modeler. Following that, the fitting of the pentameric structures to the cryo-EM map of the T=1 particle was performed manually using O (16) based on one of the pentamers from the BKV T=7 capsid. We used the VP1 pentameric model of BKV at the pentavalent position in the T=7 particles to build the atomic model of the T=1 particles. For the initial fitting, only the core of the VP1 was used, which consists of residues 16 to 296. This was based on the fact that the conformation of this pentameric structure was almost the same, with root mean-square deviations (rmsd) of less than 0.1 nm, as that in a T=7 particle.
Further, in the final stage of fitting, the position of the pentamer was optimized by the reciprocal-space rigid-body refinement between the cryo-EM map and the models. The amino acid residues 297 to 330 connecting the neighboring pentamers were omitted in this procedure. The cryo-EM map was moved into a big P1 cell (cell dimension a = b = c = 1,000 Å, α = β = γ = 90°) using the RAVE package of the Uppsala Software Factory (18) to calculate the correlation coefficient at finer grids (24). With the pentamer fivefold axis kept aligned with the icosahedral fivefold axis, the VP1 pentamer was translated along the direction of fivefold axis in 0.1 nm steps from −1.0 to +1.0 nm of the initial position, while at each step it was rotated around the fivefold axis in 1-degree intervals, from 0 to 71 degrees, relative to initial position. During the rigid-body refinement, the structure of residues 331 to 362 was moved together with the neighboring pentamer. Subsequently, the entire VP1 capsid shell was constructed at each position and orientation according to the icosahedral symmetry. The structure factors were calculated from the models with the B-factor of 100 nm2 to simulate the resolution limit of the cryo-EM reconstruction map (8, 27), and the correlation coefficient was calculated between the atomic models and the cryo-EM map using CCP4 suite (CCP4, 1994). The pentamer was moved to the position where a maximum correlation coefficient of 0.82 was reached. After the structure that connected the neighboring pentamer was refined, the correlation between the cryo-EM map described above and the final model reached a coefficient of 0.84.
RESULTS
Effect of buffer composition on VLP disassembly and the structure of reassembled particles.
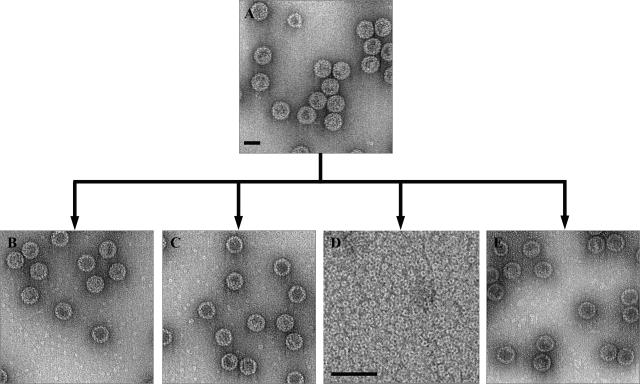
The major factors that contributed to the disassembly of T=7 VLPs into pentameric capsomers were found to include reduction of disulfide bonds and removal of calcium ions. A combination of a reducing agent and a calcium chelator was required to disassemble the particles. However, dissociation did not occur at very high ionic strength, as seen in the sample treated at ∼0.6 M NaCl, in spite of the presence of both reducing agent and EDTA (Fig. 1).
FIG. 1.
Factors guiding disassembly of BKV VLPs. (A) Electron micrograph (negative stained) of recombinant BKV VLPs (0.5 mg/ml). To find dissociation conditions, the BKV VLPs (A) were equilibrated by dialysis against different concentrations of EDTA (10 to 100 mM), 2-ME (10 to 100 mM), and NaCl (0.15 to 1 M). All experiments were done overnight at room temperature, and the resulting appearance of the samples are shown in the electron micrographs B to E (negative stain). (B) Result from dialysis against 20 mM EDTA. (C) Result from dialysis against 30 mM 2-ME. (D) Result from dialysis against 20 mM EDTA and 30 mM 2-ME. (E) Result from dialysis against 20 mM EDTA, 30 mM 2-ME, and 0.6 M NaCl. Bars, 50 nm.
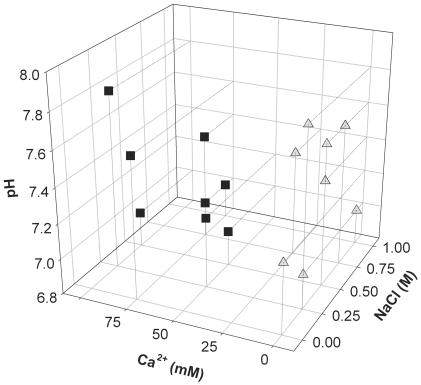
The effect of buffer composition on particle formation from free pentamers was analyzed by dialysis experiments. Samples of free capsomers were exposed to buffers containing different concentrations of CaCl2 and NaCl at various pHs (Fig. 2). Large quantities of small VLPs were formed at low calcium ion concentrations (5 to 10 mM) and in calcium-free buffers. The number of small VLPs decreased when the calcium ion concentration was increased (25 to 100 mM). Although calcium ions were needed to form the T=7 VLPs, the number of T=7 particles did not change significantly when the calcium ion concentration was increased in the 25 to 100 mM range. At above 300 mM Ca2+, VLP formation was not seen. The concentration of monovalent salt, the pH, or the ionic strength did not directly influence the reassembly of pentamers into higher oligomeric forms.
FIG. 2.
Effect of buffer composition on the shape of reassembled particles. When the VLPs had been dissociated, the sample (now containing free pentamers) was dialyzed again against buffers with different concentrations of a monovalent salt, NaCl (0.0 to 1.0 M), pH (7.0 to 7.8), and a divalent ion, Ca2+ (0.0 to 100.0 mM). Square: buffer conditions when less then 30% of the reassembled particles had a T=1 symmetry. Triangle: buffer conditions when more then 80% of the reassembled particles had a T=1 symmetry. All experiments were done at room temperature and overnight.
As mentioned above, a reducing agent was needed to disassemble the BKV VLPs, expressed in insect cells. However, the presence of 2-ME in the reassembly buffer did not prevent the formation of either small or large particles (data not shown).
Three-dimensional reconstruction of the small reassembled BKV VLP.
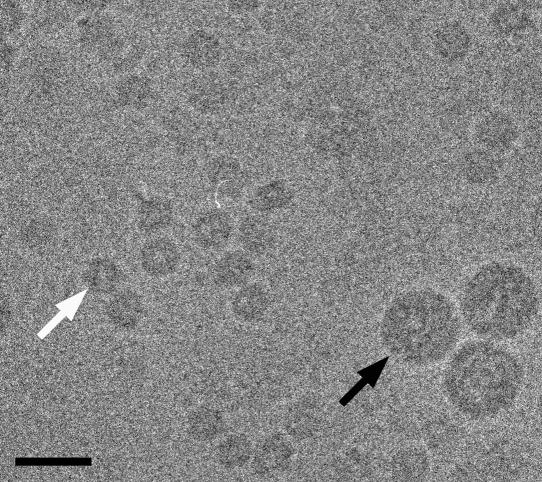
Two BKV VLPs that differed in diameter (26.4 and 50.6 nm) were observed in the low-dose micrograph (Fig. 3). The larger VLPs, with the size of the native virus, were used as an internal reference standard and showed nearly circular image profiles. That indicated that the features of the larger VLPs were well preserved in the vitrified ice. The image profile of the smaller VLPs was also circular, but they did not have the smooth surface of the larger VLPs. Before freezing, the VLP concentration was adjusted so that about 400 particles per micrograph could be selected at the magnification of 45,000.
FIG. 3.
Cryo-electron micrograph of BKV VLPs that were reassembled after overnight equilibration against 5 mM Ca2+, 150 mM NaCl, and 10 mM Tris-HCl, pH 7.4. Two populations of BKV VLPs that differed in size can be seen (white arrow, small VLP; black arrow, large native-size VLP). Bar, 50 nm.
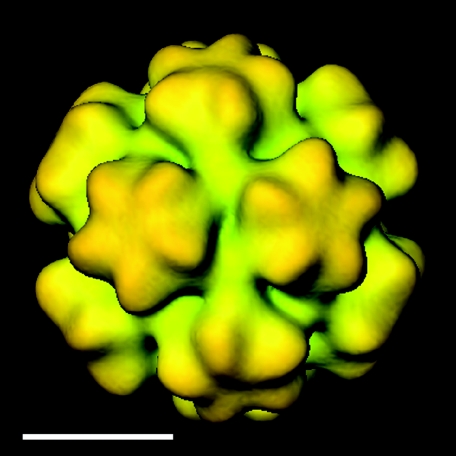
The structure of the BKV VLP was solved at ∼2.4 nm resolution from unstained, frozen-hydrated samples that were imaged with low-irradiation cryo-EM procedures. The three-dimensional reconstruction showed 12 pentameric capsomers in the smaller BKV VLP arranged according to a T=1 surface lattice placed on the 12 fivefold rotation axes of the icosahedron (Fig. 4). Due to the smaller diameter of the T=1 structure, the unique angle between the capsomers in this particle was 38° greater than that of the average angle between capsomers in the T=7 structure (21).
FIG. 4.
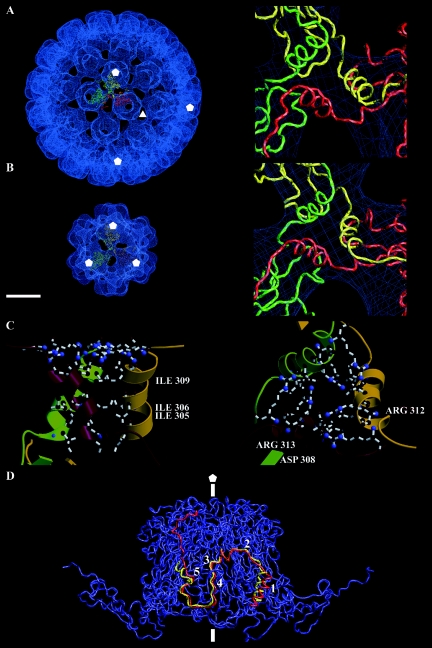
Cryo-EM three-dimensional reconstruction of the small BKV VLP from Fig. 3. The particle is viewed along the twofold axis. It has icosahedral symmetry, and the VP1 protein establishes an arrangement according to a T=1 lattice with protruding capsomer at each fivefold. Bar, 10 nm.
The morphology of the pentameric capsomers was more pronounced in the T=1 than in the T=7 structure. The T=1 capsomers had a diameter of 9.2 nm, which was slightly larger than capsomers in the T=7 structure (∼1.2 nm larger in diameter). This extra density, compared to the T=7 capsomers, was located in the outermost part of each monomer. However, the thickness of the capsid shell was similar, approximately 6.0 nm, in both the T=1 and T=7 structures.
The major intercapsomeric contact was located at the threefold axis, where the structure protruded from the bottom shell region. The interpentameric contact in the T=1 and T=7 VLPs at the threefold respective local threefold formed a Y-shaped density connecting three capsomers. This contact remained also when the contour value was raised to 2 sigma (see details in Fig. 7) or following a radial cut of the VLPs capsid shell density. The threefold contact started at a radius of 9.6 nm from the particle center and ended 11.4 nm from the particle center (21.0 to 22.4 nm in the T=7 VLP). The density in the twofold axes was much weaker than in the threefold axes. Openings in the capsid shell seen at the threefold and around the twofold axes in the T=7 structure were absent in the T=1 structure.
FIG.7.
Interpentameric interactions in the T=1 and T=7 BKV VLP. (A) Left: VP1 model of the α (yellow), α′ (red), and α′ (green) monomer fitted to the density map of the T=7 particle at the local threefold. Right: close-up of the interpentameric contact “triple-helix bundle” at the local threefold. (B) Left: three T=1 VP1 models fitted to the density map of the T=1 particle, the particle is viewed along the threefold axis. Right: close-up of the interpentameric contact found at the threefold axis in the T=1 particle. (C) Close-up of the triple-helix bundle in the smaller T=1 particle, showing possible salt-bridges and hydrophobic interactions. Right, side view; left, top view. (D) The C-terminal arm from the T=1 particle (red) and the α monomer from the T=7 particle (yellow) are here superimposed to compare interpentameric interactions. The C-terminal arms interact with two monomers in the neighboring pentamer (blue). The C-terminal arms start with a helix at the threefold (in the T=1 particle) and at the local threefold in the T=7 particle (1). The helix is connected through a long loop (2) to the J-strand (4). The two possible calcium-binding amino acids in the C-terminal arm are also marked, Glu331 (3) and Asp346 (5). The threefold and fivefold are marked as triangle and pentagons, respectively. Bar, 10 nm.
Fitting the VP1 model into the density maps of T=1 and T=7 VLPs.
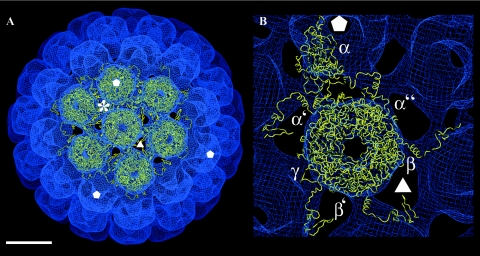
The primary sequence of BKV VP1 has a 74% similarity to the SV40 VP1 that has been solved to a 0.31 nm resolution (34). The similarity is even higher in the C-terminal arm (83%), while the main differences are in the loops at the outer surface of the monomer. A structural model of the BKV VP1 was created by aligning the primary sequence of BKV VP1 with the coordinates of VP1 from SV40 (SV40, PDB:1SVA). The model of the BKV VP1 was first fit to the density map of the T=7 particle to reveal the differences and similarities to particles with the same T-number (Fig. 5A). The only modification required to obtain an excellent fit was an adjustment for the slightly larger radius of the BKV VLP (+0.47 nm of the local sixfold pentamer and +0.58 nm of the fivefold pentamer). The six unique C-terminal arms (α, α′, and α" at the local threefold axis; β, β′ around the icosahedral threefold axis; and γ at the twofold axis) (34), responsible for the interpentameric contacts, fitted in the EM density map as well (Fig. 5B).
FIG. 5.
Fitting of the VP1 model to the density map of the larger T=7 particle structure. (A) The density map is viewed along the local sixfold axis, showing the VP1 model fitted to one fivefold pentamer and six local sixfold pentamers. Asterisk, the local threefold position. Bar, 10 nm. (B) Close-up view of the six unique monomers (α, α′, and α" at the local threefold; β, β′ around the icosahedral threefold; and γ at the twofold). The positions of the threefold and fivefold axes are marked as triangle and pentagons, respectively.
Each of the six unique monomers was tested as a possible model for the VP1 in the T=1 structure. Three of the monomers were excluded because they formed the major interpentameric contact at the twofold axis and had little or no contact at the threefold axis (not shown). In contrast, the three monomers making up the local threefold contact in the T=7 structure had their major interpentameric contacts at the threefold axis. The fivefold monomer (α) from the T=7 structure was selected as the preferable model of the three, and it was modeled in the density map of the T=1 structure.
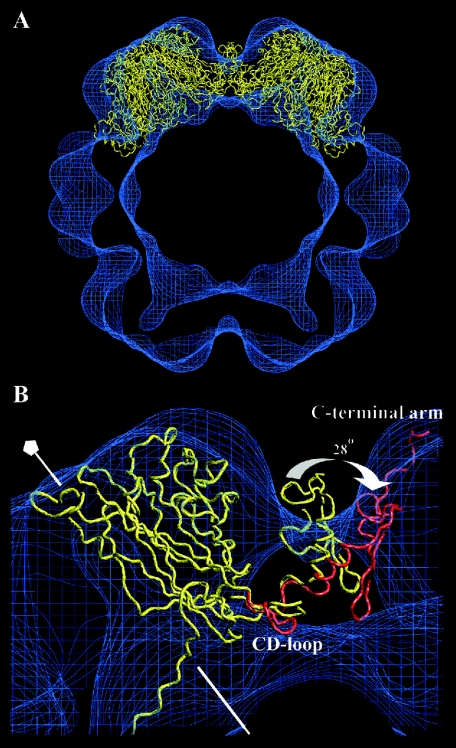
The core of the VP1 protein (residues 16 to 296) was well fitted to the EM density map (correlation coefficient of 0.82) (Fig. 6A). However, the C-terminal arm protruded out from the density map and did not reach the adjacent capsomer, as the surface curvature was higher within the small-size VLPs compared to the large-size VLPs. Therefore, the angle between the core of VP1 and the outer region of the C-terminal arm required a 28° adjustment in order for the arm to reach the adjacent capsomer (Fig. 6B). The two loops (residues 297 to 300 and 314 to 329) that linked the C-terminal helix to the VP1 protein core and the outermost region of the C-terminal arm were used to increase the angle between the protein core and the C-terminal arm. The last β-strand (the J strand) and the following loop could be modeled in good agreement to the SV40 α monomer. There were, however, additional densities at the side of the monomers. As the structure of the C-terminal domain, covering amino acid residues 348 to 362, had not yet been modeled into the T=1 particle, this domain was assumed to account for these additional densities (Fig. 6B).
FIG. 6.
Fitting of the VP1 model to the density map of the T=1 structure. (A) The fivefold monomers from the larger T=7 particle were fitted into the density map of the T=1 particle. The core of the VP1 model fitted well, while the C-terminal arm was protruding out from the density map at the threefold. This was because the angle between capsomers in the T=1 structure was 38 degrees larger than the averaged angle between capsomers in the T=7 particle. (B) Parts of two pentamers are seen from the side. There were no structural changes made in the core of the VP1 protein, except for the CD-loop. The unchanged VP1 model is shown in yellow, whereas the modulations made to fit the T=1 structure is shown in red. The fivefold axis is marked with a pentagon.
The structure of the CD loop, which consists of amino acid residues 96 to 106, was rebuilt to that of the α′ subunit to avoid a collision with the corresponding loop from an adjacent capsomer (34). This region has the highest local root-mean-square deviations between subunits in the T=7 particle, suggesting that this region is flexible (Fig. 6B). After refining the C-terminal arm and the CD-loop (the interpentameric connectors), the correlation coefficient between the cryo-EM map, described above, and the final VP1 model reached a value of 0.84.
Interpentameric contacts in the T=1 structure.
With the adjustments in the C-terminal arm, the structure of a triple-helix bundle, at the threefold axis, was similar to the interpentameric contact at the local threefold axis in the T=7 particles (amino acids 300 to 313) (Fig. 7A and B). The three helices were tightly held together by both hydrophobic interactions and salt bridges (Fig. 7C). Furthermore, in the T=1 structure the triple-helix bundle was in position to form four hydrogen bonds between each other, which were not seen in the T=7 structure. There was, however, a stronger hydrophobic effect within the helix bundle in the T=7 structure than in the T=1 structure.
To accommodate the higher curvature in the T=1 particle than that in the T=7 capsid, the C-terminal helix together with the following long loop (amino acids 314 to 329) was tilted 28° away from its own β-barrel core towards the adjacent pentamer. Through this tilt the β-strand J was able to connect with the adjacent pentamer in the T=1 structure (Fig. 7D). The long loop, together with the outermost area of the C-terminal arm, composed the region that contributed the least to the interpentameric contact in both the T=1 and the T=7 capsids. Following the loop, the J-strand made a strong interpentameric contact (with identical interactions as in the T=7 structure), starting with one (Glu331) of the two possible calcium-binding amino acids in the C-terminal arm. Amino acid 331 could, however, make an additional salt bridge with amino acid Lys195 in a nearby monomer of the adjacent pentamer. The second amino acid (Asp346) in the C-terminal arm, which was thought to be a calcium-binding site, could establish a salt bridge with amino acid Lys30.
DISCUSSION
The structure of a small VLP of the human BK polyomavirus has been studied and compared with an earlier solved BKV VLP. We have also studied the effect of the buffer composition on VLP disassembly and reassembly processes. The small BKV VLP (26.4 nm in diameter) was shown to have a T=1 surface lattice, with a pentamer located at each fivefold axis. In these T=1 particles, the main interpentameric contact was found at the icosahedral threefold axis. The angle between the pentamers was 38° greater when compared to the larger BKV VLP (50.6 nm in diameter), which has a capsid composed of 72 pentamers arranged in T=7 surface lattices (21). Our observed structure of the T=1 particle agrees very well with the computational modeling of the 12-capsomer murine polyomavirus particle done by Salunke et al. (29).
With a 74% similarity to SV40 VP1, the primary sequence of BKV VP1 was aligned with the coordinates of VP1 from SV40 (SV40, PDB:1SVA) to create a structural model of the BKV VP1 (34). This model was first fitted to the density map of the T=7 particle in order to reveal the differences and similarities to particles with the same T-number. An excellent fit of the VP1 model to the EM-density map was obtained after an adjustment of the slightly larger diameter of the BKV VLP. The six unique C-terminal arms, responsible for the interpentameric contacts, fitted in the EM density as well.
The fivefold monomer from the T=7 structure inserted well into the T=1 EM density following an adjustment to particle curvature in the hinge regions of the C-terminal arm. In T=1 particles, the pentamers were connected to each other at the icosahedral threefold axis in a similar way as at the local threefold axis in the T=7 particle. Here, seen in both particles, the C-terminal arms met in a triple-helix bundle (residues 301 to 313). In the T=1 particles this bundle appears to have four additional hydrogen bonds and to be less hydrophobic compared to the T=7 structure.
While the outermost part of the C-terminal arm (amino acids 350 to 362) appear to be the most flexible part that do not significantly contribute to the stability of either the small or large VLP, the interactions involving the J-strand is conserved. Thus, the key feature of the capsomer-intervening C-terminal arm is the ability to adjust to the pentamer's relative orientation and particle curvature. In the T=1 particle the essential adopting regions are the hinges on both side of the short C-terminal helix. The flexibility in these regions allows the J-strand to insert in the conserved orientation and the short helices from three capsomers to form a bundle, fitting with the cryo-EM map.
A similar type of flexible adaptation in the C-terminal arms has been reported for the T=1 and T=7 VLPs of the papilloma virus (7, 23). Despite lacking significant sequence similarity, the BKV VP1 (362 residues) and the human papilloma virus 16 L1 (531 residues) both form pentameric capsomers and similar architectures. Also, in the papilloma virus T=7 structure the C-terminal arm “invades” adjacent pentamers, keeping the shell together. However, in the T=1 structure the C-terminal arms exit the monomer to form the interpentameric contact at the threefold axis and then reinsert into the same monomer, making an elbow-like interpentameric contact. The glycine- and proline-rich sequence, which allows the elbow-like connection in papilloma virus, is not found in the BKV VP1 protein, nor is a similar contact fitting with the structural data available for the BKV VLPs. As a common feature, the variability in the geometry of the contacts in both the polyoma and papilloma virus is entirely due to the inherent flexibility of the C-terminal domains, while the structure of the monomer core (the pentameric ring) seems rather stable and does not change significantly between the T=1 and T=7 structures. Differing from the papilloma virus case, the T=1 and T=7 structures of the polyomavirus seem to have one interpentameric interaction in common.
We can here conclude that both Ca2+ and disulfide links are important for the stability of the recombinant BKV T=7 capsid. However, when the ionic strength of the buffer solution is increased, hydrophobic effects are more influential and keep the capsid intact also in a reducing environment, lacking calcium ions. Chen et al. (6) showed that the reducing and chelating agents worked in a specific order. The reducing agent had to be introduced in advance of the chelating agent in order for dissociation of the VLPs to occur. Thus, the disulfide bond appear to protect bound calcium ions from contact with the chelating agent. Our reassembly experiments also showed that the formation of small or large VLPs does not require the formation of disulfide bonds in vitro. This may suggest that the disulfide interaction is not necessary for the actual reassembly process but that the bond is formed after the particle has been reassembled, allowing for an increase in the stability of the VLP.
Recently, a similar study was done with SV40 VP1, where free pentamers were found to reassemble preferably into the T=1 particle in the absence of calcium (17). Contrary to our observations and to those of Salunke et al. (29), Kanesashi et al. (17) did not observe any reassembly into VLPs with buffers having physiological pH (pH 7.2) and salt concentrations (150 mM NaCl). This could be due to the use of a low, 60-μg/ml, protein concentration, compared to the 0.5- to 1-mg/ml concentration used in our and Salunke's experiments.
Both Kanesashi's and our study show that small T=1 polyoma VLPs can form without calcium, while the larger T=7 structure requires the calcium ion for assembly. Two calcium-binding sites were found in five of the six unique monomers (α, α′, α", β, and β′), while only one was found in the twofold monomer (γ) in the SV40 structure (34). One amino acid in the C-terminal arm was thought to be involved in each calcium-binding site. Even though the structure in the C-terminal arm was different among the six unique monomers, the interpentameric interactions were quite similar. The difference in structure allowed the same amino acids within the six unique monomers to have similar interpentameric interactions (hydrogen bonds, hydrophobic interactions, or salt bridges) at the three unique interpentameric contacts; at the local threefold axis, around the icosahedral threefold axis, and at the icosahedral twofold axis. However, only in the interpentameric connections at the local threefold and at the icosahedral twofold could the possible calcium-binding amino acids in the C-terminal arm form salt bridges with the adjacent pentamer. The negatively charged amino acids holding the calcium ion would repel each other in its absence. A partial explanation as to why the T=1 structure could form without calcium, whereas the T=7 structure could not, would then be that only the interpentameric contacts around the T=7, icosahedral threefold axis requires calcium for stabilization.
In conclusion, our structural study of the two BKV VLPs has shown that the triple-helix bundle is preserved in both structures and can form without Ca2+. The “flatter” contacts around the threefold axis in the T=7 particle is probably both stabilized and enforced by Ca2+, since salt bridges would not substitute in a similar conformation. The calcium concentration will in this way be the critical factor controlling the assembly of the VLPs into different sizes.
Neither of the two assembly models discussed in the literature, namely the assembly nucleus of a “five-around-one” pentamer (34) or a dimer of pentamers (4), can be deduced in our case. All the three unique contacts identified in the BKV VLPs elaborate on the formation of the J strand in the invaded pentamer. As a strong interaction, considerable energy would be needed to replace any type of unique contacts with one another. The primary contact between soluble pentamers would rely on the formation of looser contacts. These are most likely provided by the C-terminal arm at a location before the strand J forming sequence. While further investigations are needed to reveal the steps in which the polyoma VP1 pentamer assembles into particles and virions, our recent data suggests that the position of the C-terminal helix and its following tail could be stabilized through interaction with internal DNA, resulting in a more stable native-like capsid (Nilsson et al., manuscript in preparation).
Acknowledgments
We thank A. Gutierrez-Rodriguez for his fruitful discussions about protein modeling.
T. Miyamura was supported in parts by grants from the Second Term Comprehensive 10-year Strategy for Cancer Control and by the Program for Promotion of Fundamental Studies in Health Sciences of the Organization for Drug ADR Relief, R&D Promotion and Product Review of Japan (ID:01-3). R. H. Cheng was supported in parts by grants from the Swedish Research council, STINT Foundation, Tripep AB, and Foundation for Knowledge and Competence Development.
REFERENCES
- 1.Baker, T. S., J. Drak, and M. Bina. 1989. The capsid of small papova viruses contains 72 pentameric capsomers: direct evidence from cryo-EM of simian virus 40. Biophys. J. 55:243-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, T. S., and R. H. Cheng. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116:120-130. [DOI] [PubMed] [Google Scholar]
- 3.Brady, J. N., V. D. Winston, and R. A. Consigli. 1977. Dissociation of polyomavirus by the chelation of calcium ions found associated with purified virions. J. Virol. 23:717-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casini, G. L., D. Graham, D. Heine, R. L. Garcea, and D. T. Wu. 2004. In vitro papillomavirus capsid assembly analyzed by light scattering. Virology 325:320-327. [DOI] [PubMed] [Google Scholar]
- 5.Chang, D., C. Y. Fung, W. C. Ou, P. C. Chao, S. Y. Li, M. Wang, Y. L. Huang, T. Y. Tzeng, and R. T. Tsai. 1997. Self-assembly of the JC virus major capsid protein, VP1, expressed in insect cells. J. Gen. Virol. 78:1435-1439. [DOI] [PubMed] [Google Scholar]
- 6.Chen, P. L., M. Wang, W. C. Ou, C. K. Lii, L. S. Chen, and D. Chang. 2001. Disulfide bonds stabilize JC virus capsid-like structure by protecting calcium ions from chelation. FEBS Lett. 500:109-113. [DOI] [PubMed] [Google Scholar]
- 7.Chen, X. S., R. L. Garcea, I. Goldberg, G. Casini, and S. C. Harrison. 2000. Structure of small virus-like particles assembled from the L1 protein of human papillomavirus 16. Mol. Cell 5:557-567. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, R. H., V. S. Reddy, N. H. Olson, A. J. Fisher, T. S. Baker, and J. E. Johnson. 1994. Functional implications of quasi-equivalence in a T=3 icosahedral animal virus established by cryo-electron microscopy and X-ray crystallography. Structure 2:271-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frisque, R. J., G. L. Bream, and M. T. Cannella. 1984. Human polyomavirus JC virus genome. J. Virol. 51:458-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuller, S. D., S. J. Butcher, R. H. Cheng, and T. S. Baker. 1996. Three-dimensional reconstruction of icosahedral particles-the uncommon line. J. Struct. Biol. 116:48-55. [DOI] [PubMed] [Google Scholar]
- 11.Gharakhanian, E., A. K. Sajo, and M. K. Weidman. 1995. SV40 VP1 assembles into disulfide-linked postpentameric complexes in cell-free lysates. Virology 207:251-254. [DOI] [PubMed] [Google Scholar]
- 12.Griffith, J. P., D. L. Griffith, I. Rayment, W. T. Murakami, and D. L. D. Caspar. 1992. Inside polyomavirus at 25-Å resolution. Nature 355:652-654. [DOI] [PubMed] [Google Scholar]
- 13.Haynes, I. I., J. I. Chang, and R. A. Consigli. 1993. Mutations in the putative calcium-binding domain of polyomavirus VP1 affect capsid assembly. J. Virol. 67:2486-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishizu, K. I., H. Watanabe, S. I. Han, S. N. Kanesashi, M. Hoque, H. Yajima, K. Kataoka, and H. Hand. 2001. Roles of disulfide linkage and calcium ion-mediated interactions in assembly and disassembly of virus-like particles composed of simian virus 40 VP1 capsid protein. J. Virol. 75:61-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jao, C. C., M. K. Weidman, A. R. Perez, and E. Gharakhanian. 1999. Cys9, Cys104 and Cys207 of simian virus 40 VP1 are essential for inter-pentamer disulfide-linkage and stabilization in cell-free lysates. J. Gen. Virol. 80:2481-2489. [DOI] [PubMed] [Google Scholar]
- 16.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 17.Kanesashi, S. N., K. I. Ishizu, M. A. Kawano, S. I. Han, S. Tomita, H. Watanabe, K. Kataoka, and H. Handa. 2003. Simian virus 40 VP1 capsid protein forms polymorphic assemblies in vitro. J. Gen. Virol. 84:1899-1905. [DOI] [PubMed] [Google Scholar]
- 18.Kleywegt, G. J., J. Y. Zou, M. Kjeldgaard, and T. A. Jones. 2001. Around O. In M. G. Rossmann and E. Arnold (ed.), International tables for crystallography, vol. F. Crystallography of biological macromolecules, p. 353-356. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 19.Li, P. P., A. Nakanishi, S. W. Clark, and H. Kasamatsu. 2002. Formation of transitory intrachain and interchain disulfide bonds accompanies the folding and oligomerization of simian virus 40 vp1 in the cytoplasm. Proc. Natl. Acad. Sci. USA 99:1353-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, P. P., A. Nakanishi, M. A. Tran, K. I. Ishizu, M. Kawano, M. Phillips, H. Handa, R. C. Liddington, and H. Kasamatsu. 2003. Importance of VP1 calcium-binding residues in assembly, cell entry, and nuclear entry of SV40. J. Virol. 77:7527-7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, T. C., N. Takeda, K. Kato, J. Nilsson, L. Xing, L. Haag, R. H. Cheng, and T. Miyamura. 2003. Characterization of self-assembled virus-like particles of human polyomavirus BK generated by recombinant baculoviruses. Virology 311:115-124. [DOI] [PubMed] [Google Scholar]
- 22.Liddington, R. C., Y. Yan, J. Moulai, R. Sahli, T. L. Benjamin, and S. C. Harrison. 1991. Structure of simian virus 40 at 3.8-A resolution. Nature 354:278-284. [DOI] [PubMed] [Google Scholar]
- 23.Modis, Y., B. L. Trus, and S. C. Harrison. 2002. Atomic model of the papillomavirus capsid. EMBO J. 21:4754-4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Navaza, J. 1994. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A 50:157-163. [Google Scholar]
- 25.Ou, W. C., L. H. Chen, M. Wang, T. H. Hseu, and D. Chang. 2001. Analysis of minimal sequences on JC virus VP1 required for capsid assembly. J. Neurol. Virol. 7:298-301. [DOI] [PubMed] [Google Scholar]
- 26.Rodgers, R. E. D., D. Chang, X. Cai, and R. A. Consigli.1994. Purification of recombinant budgerigar fledgling disease virus VP1 capsid protein and its ability for in vitro capsid assembly. J. Virol. 68:3386-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossmann, M. G. 2000. Fitting atomic models into electron-microscopy maps. Acta Crystallogr. D 56:1341-1349. [DOI] [PubMed] [Google Scholar]
- 28.Salunke, D. M., D. L. Caspar, and R. L. Garcea. 1986. Self-assembly of purified polyomavirus capsid protein VP1. Cell 46:895-904. [DOI] [PubMed] [Google Scholar]
- 29.Salunke, D. M., D. L. D. Caspar, and R. L. Garcea. 1989. Polymorphism in the assembly of polyomavirus capsid protein VP1. Biophys. J. 56:887-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt, U., R. Rudolph, and G. Böhm. 2000. Mechanism of assembly of recombinant murine polyomavirus-like particles. J. Virol. 74:1658-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: and automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sedzik, J. 1994. DESIGN -A guide to protein crystallization experiments. Arch. Biochem. Biophys. 308:342-348. [DOI] [PubMed] [Google Scholar]
- 33.Stehele, T., Y. Yan, T. L. Benjamin, and S. C. Harrison. 1994. Structure of murine polyomavirus complexed with an oligosaccharide receptor fragment. Nature 369:160-163. [DOI] [PubMed] [Google Scholar]
- 34.Stehle, T., S. J. Gamblin, Y. Yan, and S. C. Harrison. 1996. The structure of simian virus 40 refined at 3.1 Å resolution. Structure 4:165-182. [DOI] [PubMed] [Google Scholar]
- 35.Walker, D. L., and R. J. Frisque. 1986. The biology and molecular biology of JC virus, p. 161-193. In N. P. Salzman (ed.) The papovaviridae, vol. 1. Plenum Press, New York, N.Y.
- 36.Walter, G., and W. Deppert. 1975. Intermolecular disulfide bonds: an important structural feature of the polyomavirus capsid. Cold Spring Harbor Symp. Quant. Biol. 39:255-257. [DOI] [PubMed] [Google Scholar]
- 37.Wickham, T. J., and G. R. Nemerow. 1993. Optimization of growth methods and recombinant protein production in BTI-Tn-5B1-4 insect cells using the baculovirus expression system. Biotechnol. Prog. 9:25-30. [DOI] [PubMed] [Google Scholar]
- 38.Xing, L., K. Kato, T. Li, N. Takeda, T. Miyamura, L. Hammar, and R. H. Cheng. 1999. Recombinant hepatitis E capsid protein self-assembles into dual-domain T=1 particle presenting native virus epitopes. Virology 265:35-45. [DOI] [PubMed] [Google Scholar]
- 39.Xing, L., K. Tjärnlund, B. Lindqvist, G. G. Kaplan, D. Feigelsto, R. H. Cheng, and J. Casanovas. 2000. Distinct cellular receptor interactions in poliovirus and rhinoviruses. EMBO J. 19:5081-5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan, Y., T. Stehle, R. C. Liddington, H. Zhao, and S. C. Harrison. 1996. Structure determination of simian virus 40 and murine polyomavirus by a combination of 30-fold and 5-fold electron-density averaging. Structure 4:157-164. [DOI] [PubMed] [Google Scholar]