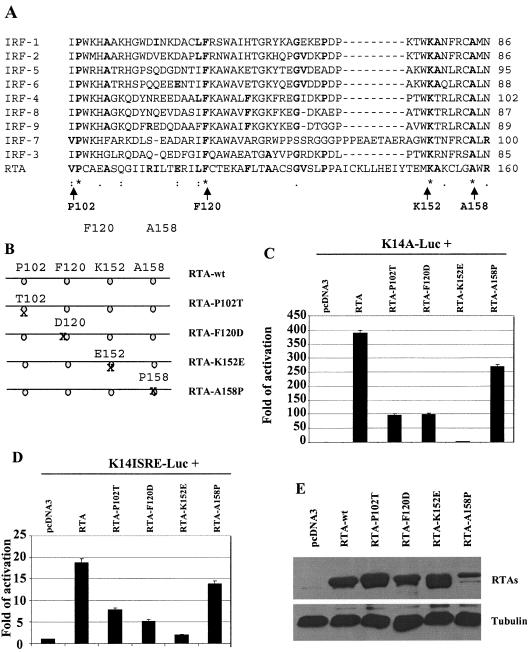
FIG. 4.
Mutations in the DNA-binding domain of RTA affect activation of the K14-ORF74 promoter. A. RTA has certain similarities to IRF proteins. The Clustal W program was used for the alignment of RTA with all human IRF members. Gaps were introduced to allow best alignment. The identity and coordinates of mutated amino acids in RTA are shown at the bottom with arrows. B. Schematic diagram of RTA mutant constructs. The coordinates and identities of the amino acids are shown. The positions of amino acids are not on scale. C. Mutations in the DNA-binding domain of RTA reduce activation of the K14-ORF74 promoter. 293T cells were transfected with reporter construct, K14A-luc, along with CMV-β-Gal and equal amounts (50 ng) of RTA or various mutant expression plasmids. The reporter activity is expressed relative to the vector control. Luciferase activity was normalized by β-galactosidase activity. Standard deviations are shown. D. Mutations in the DNA-binding domain of RTA reduce activation of K14-ISRE. 293T cells were transfected with reporter construct, K14ISRE-luc, along with CMV-β-Gal and equal amounts (0.3 μg) of RTA or various mutant expression plasmids. The reporter activity is expressed relative to the vector control. Standard deviations are shown. E. Expression levels of RTA mutants. Western blotting with RTA as well as tubulin antibodies was performed. The cell lysates from transfected cells shown in panels C or D are indicated. The identities of proteins are shown.