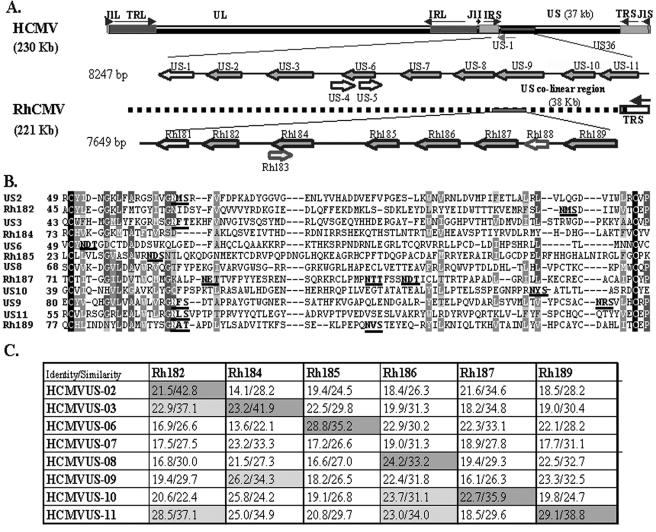
FIG. 1.
The US6 family of HCMV and RhCMV. (A) Schematic representation of HCMV and RhCMV genomes depicting the relative locations and orientations of US6 family members in the genome of HCMV and their homologues in RhCMV. US6 family genes are shown in dark gray. (B) Multiple sequence alignment (ClustalW) of the predicted Ig folds of all US6 family members of HCMV and RhCMV. Identical residues are shown as white on black background. Similar residues are shown as white on gray background. Conserved residues are represented as light gray on gray background, and weakly similar residues are dark gray on a light gray background. The N-linked glycosylation sites are in boldface letters and are underlined. (C) Pairwise sequence alignment between US6 family ORFs of HCMV and RhCMV. The percent similarity and identity were calculated by GAP alignment (PAM120 amino acid substitution matrix) with a gap penalty of 14 and an extension penalty of 2.