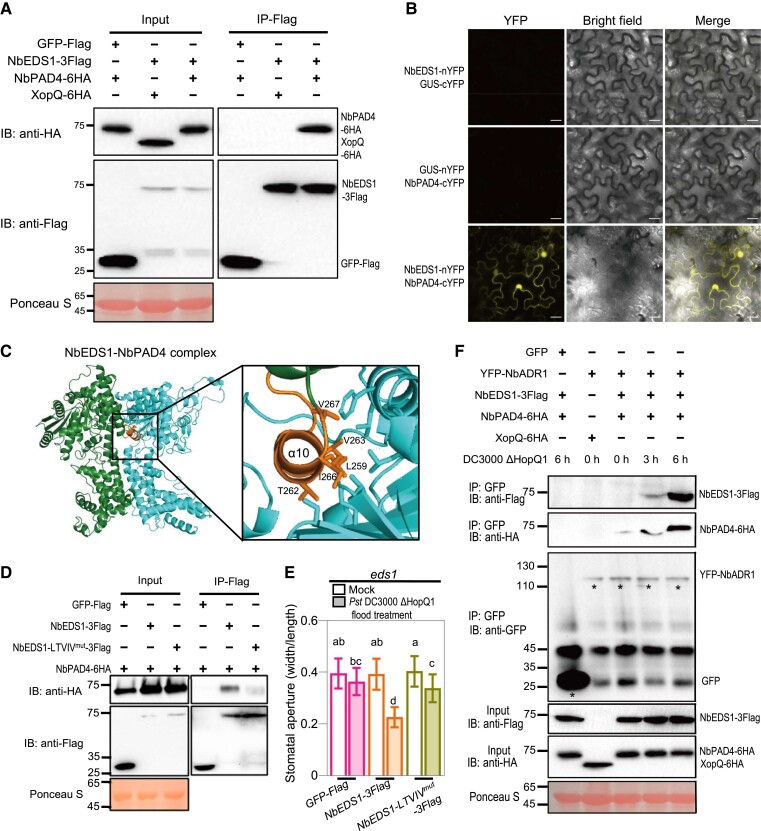
Figure 5.
NbADR1 physically associates with the preformed NbPAD4-NbEDS1 complex upon Pst DC3000 ΔHopQ1 infection. A) Co-IP assay showed the interaction of NbEDS1 with NbPAD4. The NbPAD4-6HA or XopQ-6HA (as a negative control) was transiently coexpressed with NbEDS1-3flag or the GFP-flag control in Nicotiana benthamiana leaves. The OD600 for each Agrobacterium was adjusted to 0.3. The total proteins were immunoprecipitated with the anti-Flag agarose beads, and the IP product proteins were detected by immunoblotting using the anti-flag or anti-HA antibody. Ponceau-S staining of Rubisco was used as a loading control. B) BiFC assay showed the interactions of NbEDS1 with NbPAD4 in epidermal cells of N. benthamiana leaves. The N-terminal fragment of yellow fluorescent protein (nYFP)-fused NbEDS1, the C-terminal fragment of YFP (cYFP)-fused NbPAD4, and the controls β-glucuronidase (GUS)-nYFP and GUS-cYFP were transiently coexpressed in N. benthamiana leaves, respectively. The OD600 for each Agrobacterium was adjusted to 0.3. YFP fluorescence was detected under the confocal microscope with identical gain settings (laser, 514 nm, 6%; collection bandwidth, 526 to 588 nm; pinhole, 1.20 AU; master gain, 660.0) at 30 h after coexpression. Scale bars represent 20 μm. C) The homology model of the NbEDS1-NbPAD4 protein complex based on the structure of the Arabidopsis AtEDS1-AtPAD4 complex. The key residues predicted for NbEDS1-NbPAD4 interface were marked with orange in NbEDS1. D) Co-IP assay showed that the interaction of NbEDS1 with NbPAD4 is largely disrupted by the “LTVIV” mutation (L259E, T262F, V263E, I266E, and V267E) within NbEDS1. The OD600 for each Agrobacterium was adjusted to 0.3. The total proteins were immunoprecipitated with the anti-Flag agarose beads, and the IP product proteins were detected by immunoblotting using the anti-flag or anti-HA antibody. Ponceau-S staining of Rubisco was used as a loading control. E) Stomatal apertures in leaves of N. benthamiana eds1 with transient expression of GFP-Flag, NbEDS1-3Flag, or NbEDS1-LTVIV-3Flag, after 1 h of flood treatment with mock (10 mM MgCl2) or Pseudomonas syringae pv. tomato (Pst) DC3000 ΔHopQ1 (OD600 = 0.4). The OD600 for each Agrobacterium was adjusted to 0.5. Data are means ± Sd; n = 50 stomata. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). F) Co-IP assay showed that NbADR1 recruits NbPAD4 and NbEDS1 upon Pst DC3000 ΔHopQ1 spray infection. The YFP-NbADR1 and GFP were transiently coexpressed with XopQ-6HA, NbEDS1-3Flag, or NbPAD4-6HA variants in N. benthamiana leaves, respectively, and the leaves were subsequently inoculated with Pst DC3000 ΔHopQ1 (OD600 = 0.4). The OD600 for each Agrobacterium was adjusted to 0.3 (except that for YFP-NbADR1 was 0.6). The total proteins were extracted from the leaves at 0, 3, or 6 h after treatment and were immunoprecipitated with the anti-GFP agarose beads. The IP product proteins were detected by immunoblotting using the anti-Flag or anti-HA antibody. Ponceau-S staining of Rubisco served as a loading control. Asterisks indicate the bands of GFP or YFP-ADR1. All these experiments were repeated 3 times with similar results. IB, immunoblotting; IP, immunoprecipitation.