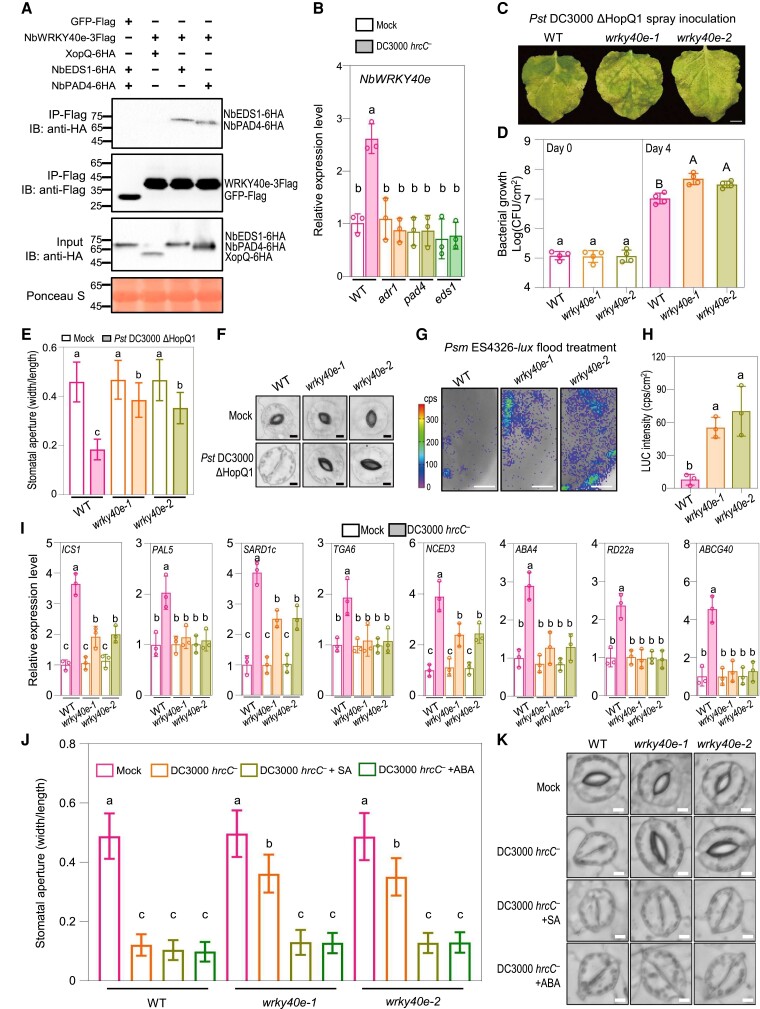
Figure 6.
NbWRKY40e associates with NbEDS1 and NbPAD4 and mediates stomatal immunity. A) Co-IP assay showed that NbWRKY40e associates with NbEDS1 and NbPAD4. The NbEDS1-6HA, NbPAD4-6HA, or XopQ-6HA was transiently coexpressed with NbWRKY40e-3flag or the GFP-flag control in Nicotiana benthamiana leaves, respectively. The OD600 for each Agrobacterium was adjusted to 0.3. Total proteins were immunoprecipitated with the anti-Flag agarose beads, and the IP product proteins were detected by immunoblotting using the anti-flag or anti-HA antibody. Ponceau-S staining of Rubisco was used as a loading control. B) Quantitative real-time PCR analysis of NbWRKY40e in N. benthamiana wild-type (WT) and the indicated mutants at 2 h postspray inoculation with mock (10 mM MgCl2) or Pseudomonas syringae pv. tomato (Pst) DC3000 hrcC− (OD600 = 0.4). Data are means (±Sd) of 3 biological replicates from independent plants. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). C, D) Disease symptoms C) and bacterial populations D) in leaves of indicated plants at 6 d C), or 0 and 4 d D) postspray inoculation with Pst DC3000 ΔHopQ1 (OD600 = 0.4). Data are means (±Sd) of 4 biological replicates from independent plants. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). Scale bars represent 1 cm. E, F) Stomatal apertures E) and images of stomata F) in leaves of N. benthamiana WT, wrky40e-1 and wrky40e-2 after 1 h of flood treatment with mock (10 mM MgCl2) or Pst DC3000 ΔHopQ1 (OD600 = 0.4). Data are means ± Sd; n = 50. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). Scale bars represent 5 μm. G, H) Bacterial pathogen entry assay in leaves of N. benthamiana WT and the indicated wrky40e mutants. The leaves were photographed and quantified by a CCD imaging system after 1 h of Psm ES4326-lux flood treatment (OD600 = 0.5). Data are means (± Sd) of 3 biological replicates from 3 independent plants. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). Scale bars represent 1 cm. I) Quantitative real-time PCR analysis of the representative SA and ABA pathway genes in N. benthamiana WT and the indicated wrky40e mutants at 2 h postspray inoculation with mock (10 mM MgCl2) or Pst DC3000 hrcC− (OD600 = 0.4). Data are means (±Sd) of 3 biological replicates from independent plants. Letters indicate significant differences by one-way ANOVA analysis (Tukey's post hoc test, P < 0.05). J, K) Stomatal apertures J) and images of stomata K) in leaves of N. benthamiana WT and the indicated wrky40e mutants after 1 h of flood treatment with mock (10 mM MgCl2), Pst DC3000 hrcC− (OD600 = 0.4), or Pst DC3000 hrcC−(OD600 = 0.4) plus SA (the form of sodium salicylate) or ABA, respectively. Data are means ± Sd; n = 50 stomata. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). Scale bars represent 5 μm. The experiments were repeated 3 times with similar results. IB, immunoblotting; IP, immunoprecipitation; CFU, colony-forming unit; cps, counts per second.