Abstract
Propagation of E1 region replacement adenovirus vectors in 293 cells results in the rare appearance of replication-competent adenovirus (RCA). The RCA genome contains E1 DNA acquired from the 293 cellular genome. The Luria-Delbrück fluctuation test was adapted to measure RCA formation rates. To test if structure affected rate, we measured rates during the production of adenovirus vectors with genomes containing three different expression cassette arrangements. The vectors had different extents of sequence identity with integrated Ad5 DNA of 293 cells and had different distributions of identity flanking the expression cassettes. Empty cassette vector RCA rates ranged from 2.5 × 10−8 to 5.6 × 10−10. The extent of sequence identity was not an accurate RCA rate predictor. The vector with the highest RCA rate also had the least overall sequence identity. To define factors controlling RCA generation, adenovirus vectors expressing E2F family proteins, known to modulate recombination gene expression, and overexpressing the human Rad51 recombination protein were analyzed. Compared to their corresponding empty vectors, RCA rates were not increased but were slightly decreased. Initial results suggested expression cassette orientation and/or transcription direction as potential RCA rate modifiers. Testing adenovirus vectors with identical transgene cassettes oriented in opposite directions suggested that transcription direction was not the basis of these rate differences. Thus, the overall structure and location of the transgene cassette had the largest effect on RCA rate. The RCA fluctuation test should be useful for investigators who require accurate measurements of targeted recombination and the probability of RCA formation during stock production.
Recombination within uninfected mammalian cells is induced by double-strand DNA breaks (37) which are repaired by homologous recombination (HR) or by nonhomologous end joining (NHEJ) (reviewed in references 39 and 50). Within infected cells, recombination of adenovirus DNA with itself or with host cell DNA leads to a number of outcomes. HR mechanisms are responsible for DNA exchanges between freely replicating viral genomes (9, 51), adenoviral genome evolution (28, 38), targeted DNA exchanges with the cellular genome (6), and the appearance of replication-competent adenoviruses (RCA) during production of E1-deleted or E1-mutated adenovirus stocks in 293 cells (14, 22, 54). Recombination by nonhomologous mechanisms results in adenovirus-induced cell transformation (7, 26; reviewed in reference 8) and genome concatemerization in cells infected by E4-deleted adenoviruses (48).
In theory, adenovirus DNA recombination is subjected to all of the recombination mechanisms of the host cell. This idea has sparked investigations into complex host cell recombination mechanisms using adenovirus as a simple and easily manipulated tool (reviewed in reference 53). In practice, the simplicity of this idea is complicated by the fact that adenovirus proteins alter at least some host cell recombination mechanisms (4, 43, 49). Determining how cellular recombination pathways act upon adenovirus genomes and how adenovirus-encoded proteins modify host recombination mechanisms will contribute to a better understanding of adenovirus replication and host cell biology.
Many single-stranded and double-stranded adenovirus genomes are produced during a productive infection (12, 44; early references are reviewed in reference 19), providing ample targets to the host cell's HR machinery. It is not presently known if HR is modified during adenovirus infection. It is known, however, that small increases in cellular Rad51 levels can increase HR (45). Increased levels of E2F1 result in higher levels of Rad51 mRNA (42). It is possible that Rad51 expression is increased after wild-type adenovirus infection as a result of the increased E2F1 transcription factor activity caused by E1A expression (20). Furthermore, adenovirus can activate signaling protein kinases that control both HR and NHEJ after DNA strand breaks (4).
Cellular proteins of infected cells recognize the ends of the linear adenovirus genomes and, as inferred from the results of Weiden and Ginsberg, will attempt NHEJ to ligate individual genomes into concatemers (48). Adenovirus produces two proteins, encoded by E4 ORF3 and E4 ORF6, which inhibit the NEHJ pathways and concatemer formation but continue to allow homologous DNA recombination (2, 10, 30, 33). The exact host recombination pathways used during adenovirus infection remain undefined, but they are likely associated with or modified by adenovirus DNA replication, the high number of single-stranded and double-stranded adenovirus genomes produced during a productive infection, and/or virus-encoded proteins.
Even though the molecular mechanisms of adenovirus DNA recombination remain unknown, recombination frequencies have been determined. HR between freely replicating adenovirus genomes (9, 51, 55, 57) is a relatively frequent event, as is that of replicating adenovirus DNA with the covalently integrated adenovirus DNA of rat F4 cells (6). The occurrences of both of these events are high enough to be expressed in percentages. In contrast, the targeted recombination frequencies observed when growing E1-deleted adenoviruses in 293 cells, using the acquisition of E1A DNA and generation of RCA as markers for HR, are extremely low (<10−8) and subject to stock-to-stock fluctuation (14, 22). This is even lower than the reported frequency (10−3 to 10−5 per cell) of nontargeted recombination between replication-deficient E1-substituted adenovirus DNA and cellular DNA of several different human and rodent cells (13, 29).
By definition, RCA will replicate in permissive host cells lacking complementing adenovirus gene products. RCA contamination of adenovirus vector stocks is a practical concern in human gene therapy and experimental biology. For clinical purposes, the U.S. Food and Drug Administration requires that there be less than 1 RCA in 3 × 1010 viral particles (Food and Drug Administration, BRMAC Meeting 30: adenovirus titer measurements and RCA levels, 5 April 2001. Food and Drug Administration, Rockville, Md.). Because up to 1013 virus particles can be injected per patient, injections will contain some RCA. RCA occurs because production stocks of adenovirus vectors are made using 293 cells. The 293 cell genome contains Ad5 DNA sequences spanning nucleotides 1 to 4344 (23) that provide a recombination target to E1-deleted adenovirus vectors. Because the frequency of RCA is low, most stocks pass the Food and Drug Administration requirement. Unfortunately, the more often an E1 deletion mutant is serially passaged through 293 cells, the easier it becomes to detect RCA in the viral yield (22).
RCA frequency is a proportional measurement. The frequency varies among multiple viral stocks produced from a single parental stock. This scatter reflects the time RCA first appear in the viral replication cycle. Frequency estimates are skewed further if there are differential growth rates of RCA compared to its progenitor. Thus, RCA frequencies are only general estimates of the probability of RCA formation during stock generation.
To more accurately define the chance of having RCA in an adenovirus stock, we used a modification of the Luria-Delbrück fluctuation test (24). This test measures RCA rate. Moreover, by performing a simultaneous RCA analysis on a parallel set of viral progeny populations, this test excludes the variances described above. The RCA fluctuation test proved valuable in comparative studies designed to determine if adenovirus vector structure or transgene expression modified RCA susceptibility. Using this approach, we determined the rates of RCA formation of several adenovirus vectors whose E1 regions had three different amounts and arrangements of sequence identity with the integrated adenovirus DNA of 293 cells; if leftward or rightward transcription of a transgene expression cassette influenced rate; and if overexpression of proteins involved, directly or indirectly, in cellular HR affected the RCA rate. The data generated are important in the study of targeted recombination events in 293 cells and also provide a baseline for experiments aimed at defining host cell proteins directly involved in the process.
MATERIALS AND METHODS
Cell lines.
293A cells were purchased from Quantum Biotechnologies, Inc. (Montreal, Quebec, Canada). A549 and 293 cells were gifts from H. S. Ginsberg and Eric Frost, respectively. Cells were grown in Dulbecco's modified Eagle medium (Mediatech, Inc., Herndon, Va.) supplemented with 10% FetalClone II (HyClone, Logan, Utah), 100 U/ml penicillin, and 100 μg/ml streptomycin (Mediatech, Herndon, Va.). Cells were incubated in a 37°C humidified incubator with 5% CO2 in air.
Virus construction and stock production.
Transgenes were cloned into expression cassettes of pACE (56) or pDC516 (32) shuttle vectors. Transcription from these cassettes was controlled by the human (Hs) or mouse (Mm) CMV IE enhancer/promoter, respectively. For the construction of Ad.HisSUMO-1, the entire SV40-based expression cassette was cloned from pSG5-HisSUMO-1 in both orientations into pDC511 (32), which lacks an expression cassette. Viruses were constructed by methods based on HR or site-specific recombination of two plasmids. In the first system that relied on HR to produce adenovirus vectors, pACE DNA was transfected into 293A cells with pJM17 DNA (27). In the second system that used frt site-specific recombination to generate adenovirus vectors, pDC516 DNA or pDC511 DNA was transfected into 293A cells with pBHG,frt,ΔE1,E3,FLP (32). Viruses were grown and plaque purified using 293A cells. Plaque-initiated stocks were crude lysates with the exceptions of Ad.HsRad51 and Ad.HsCMV, which were further purified using CsCl gradients.
RCA frequency analysis.
Stocks that were generated using 293 cells were titrated using a 293 cell-based fluorescent focus assay (34). To determine the number of RCA per ml, 60-mm-diameter dishes of A549 cells were inoculated with 0.2-ml dilutions of the viral stocks for a period of 2 h with intermittent shaking. After adsorption, a standard plaque assay was performed, and plaques were enumerated at day 13 after cells were stained with neutral red (56). To calculate RCA frequency, the number of RCA per ml were divided by the total number of viruses per ml in the 293 cell-derived stock.
Fluctuation tests.
Twelve or 13 35-mm plates of 293 cells were infected with adenovirus vectors using 100 to 1,000 focus-forming units (FFU)/plate. Adsorption was done over a period of 2 h in a volume of 0.2 ml per plate, 2.5 ml of infecting fluid (21) was added, and cells were incubated at 37°C and examined daily. After all cells displayed CPE, they were subjected to freeze-thawing to produce the “original yield.” The original yield was assayed as follows: 0.1 ml was taken to determine the titers by fluorescent focus assay on 293 cells, 0.2 ml was used to infect A549 cells grown in 35-mm-diameter plates, and the remainder was used to infect A549 cells grown in 150-mm-diameter plates. The choice of plate sizes was arbitrary. A large enough number of A549 cells had to be used to avoid “input” cytopathic effect (CPE). In stocks with low amounts of RCA or stocks that lacked RCA, duplicate assays also provided greater testing confidence. Infected A549 cells were incubated at 37°C in Dulbecco's modified Eagle medium supplemented with 2% fetal bovine serum and the antibiotics listed above. After 10 to 13 days of growth in A549 cells, the cells were subjected to three cycles of freeze-thawing and 0.3 ml per sample was used to infect fresh A549 cells grown in 35-mm-diameter plates (passage 2). After 10 to 13 days, the passage 2-infected cells were subjected to freeze-thawing and 0.3 ml was used to infect fresh 35-mm-diameter plates of A549 cells. An original yield was scored positive when A549 cells displayed CPE at any step of serial infection. Apparent RCA were verified by molecular methods.
The modified Luria-Delbrück (24) formula to determine the number of RCA per virus per average yield is as follows: RCA rate = −ln(number of original yields without RCA/total number of original yields)/average yield per plate. The average yield per plate was the average titer per plate, defined in FFU/ml, in a fluctuation set multiplied by the volume (2.5 ml) of the original yield.
DNA analysis.
RCA DNA was prepared from infected A549 cells by a modified Hirt DNA extraction protocol (15, 47). HindIII digestion of extracted DNA was performed, samples were size fractionated through 1.0% agarose gels, and bands were visualized by transillumination using UV light. Southern transfer-hybridization analysis was performed on a subset of samples using E1A or E1B DNA as a probe to verify RCA DNA. Agarose gel electrophoresis of PCR products, produced using primers designed by Lochmuller (22), was done to verify RCA DNA from A549 cells infected by a CPE-inducing stock of Ad.HisSUMO-1 (right [R] to left [L]).
RESULTS
Frequencies of RCA in adenovirus vector stocks.
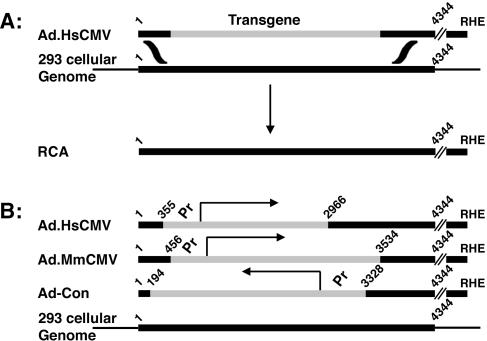
RCA are generated by an HR mechanism that is thought to involve a double crossover event in which the freely replicating vector genome acquires wild-type DNA from the 293 cell genome (Fig. 1A). The vector transgene cassette is replaced by functional Ad5 E1A and E1B DNA. RCA can be identified using bioassays, which test viral growth in cells lacking adenovirus E1A and E1B, or by molecular methods, such as restriction endonuclease mapping, Southern blot analysis, or PCR. By using a bioassay that detected RCA growth on A549 cells, we determined the levels of RCA in stocks of passaged viral populations and in stocks started using plaque-purified adenovirus vectors. The structures of the RCA genomes were verified by molecular methods.
FIG. 1.
(A) Model of RCA generation in 293 cells. The integrated DNA of 293 cells replaces the expression cassette of adenovirus vectors by an HR-based double crossover event. Theoretically, the crossover events can occur at any position with sequence identity. (B) Adenovirus vector structures and sequence identity distributions relative to the integrated Ad5 DNA of 293 cells. Black rectangles represent adenovirus DNA. Black lines represent 293 cellular DNA. Arrows represent transcriptional direction. Gray rectangles represent expression cassette DNA. Ad.HisSUMO-1 (L to R) and Ad.HisSUMO-1 (R to L) sequence identity distributions are the same as those of Ad.MmCMV. RHE, right-hand end.
Members of the E2F transcription factor family coordinate the expression of genes in a cell cycle-dependent manner (reviewed in reference 3). Overexpressing members of the E2F family can also increase or decrease the expression of genes controlling recombination (25, 35, 36). To test if they affected RCA production, we obtained adenovirus vectors that produced high levels of E2F family members (Ad.E2F1, Ad.E2F2, Ad.E2F3, AdE2F4, Ad.E2F5, and Ad.DP1). New stocks were produced using 293 cells and tested for the presence of RCA using A549 cells. RCA were present in each of our stocks.
To separate the desired vectors from RCA, viruses were plaque purified. Plaque-initiated stocks also contained RCA. RCA were enumerated by plaque assay on A549 cell monolayers. Suspected RCA plaques were isolated, and viral stocks were produced. Restriction endonuclease mapping and Southern blot analyses of the RCA genomes were consistent with replacement of transgene cassettes with 293 cell-derived E1 DNA. The frequency of RCA in the Ad.E2F1 stock was 0.2 × 10−6 and ranged from 1.6 × 10−6 to 6.8 × 10−6 in Ad.E2F2, Ad.E2F3, Ad.E2F4, Ad.E2F5, and Ad.DP-1 plaque-initiated stocks. The amount of RCA in the E2F1 stock was lower than that of all other E2F transgene-containing stocks, which either reflected a randomness of the assay or showed that overexpression of E2F1 decreased the activity of the RCA-generating recombination machinery.
In contrast to the above findings, plaque-initiated passage 1 or 2 stocks of E1 region-replacement adenovirus vectors constructed in our laboratory contained significantly lower levels of RCA. These vectors contained and highly expressed a variety of transgenes implicated in DNA recombination, DNA repair, or the cancer cell phenotype. The genomes of these vectors (Fig. 1B) were different from those used in the production of the Ad.E2F series and Ad.DP1. These results raised the possibility that viral genome structure, the direction of transcription, or overexpression of the transgene-encoded protein influenced the generation of RCA.
Fluctuation test to determine the rate of RCA generation in 293 cells.
To better understand the determinants of RCA formation, it was necessary to devise a system that avoids the random fluctuations seen when the amount of RCA in multiple stocks of a given vector is measured. With this goal in mind, the Luria-Delbrück fluctuation test was adapted to determine the rates of RCA formation.
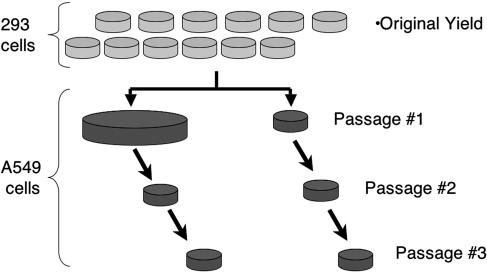
The experimental protocol for RCA rate analysis is depicted in Fig. 2. A series of 12 35-mm-diameter plates of 293 cells were infected with 100 to 1,000 FFU of each plaque-purified vector per plate. Based on prior RCA frequency measurements, this amount of virus was expected to be RCA free. The infected cells were incubated until all cells displayed advanced CPE. The cells were subjected to freeze-thawing to release the virus designated the “original yield.” The total viral yield per plate was determined by fluorescent focus assay using 293 cells. RCA detection was based on the biological endpoint of CPE in A549 cells. Two different-sized plates of A549 cells were used for the initial RCA analyses so that we could assay most (>95%) of the original yield and to provide replicates for RCA detection. Original yields were scored as negative for the presence of RCA after three sequential CPE-free passages on A549 cells. The presence of RCA in the “CPE-positive” yields from the A549 cells was confirmed by restriction endonuclease mapping of viral DNA. The acquisition of E1 DNA was confirmed by Southern blot analysis using either E1A- or E1B-specific probes.
FIG. 2.
Fluctuation test protocol for RCA rate determination. 293 cells were infected using 100 to 1,000 FFU per plate of each adenovirus vector. See Materials and Methods for additional information.
Rate of RCA generation in 293 cells: importance of vector genome organization.
To address the relationships between RCA rates and overall sequence identity or identity distributions among vector genomes and the integrated Ad5 DNA of 293 cells, we evaluated three structurally different vectors with empty or occupied transgene cassettes (Fig. 1B). The vectors with the greatest overall sequence identity are based on Ad.HsCMV. It has the largest stretch of sequence identity on the right side of the expression cassette and an intermediate amount of sequence identity on the left side of the cassette. Ad-Con and Ad.MmCMV share 1,210 or 1,266 bp of sequence identity with the integrated adenovirus DNA of 293 cells, respectively, but the distributions of their identities differ. The results of fluctuation analyses of Ad-Con and Ad.MmCMV are listed in Table 1. The comparative experiments were done using two sets of 12 original yields per virus. Each set was produced on a different day. Although approximately 16-fold more viruses were produced per plate using Ad.MmCMV than using Ad-Con, a smaller percentage of Ad.MmCMV-independent yields (3.25-fold fewer) contained RCA. The rates of RCA formation for these viruses are summarized in Table 4 and show that there was a significant increase in the RCA rate when producing stocks of Ad-Con. In contrast to the idea that more overall virus:293 cell sequence identity would result in a higher rate of RCA formation, Ad-Con RCA formation rate was greater than that of either Ad.MmCMV (Table 4) or Ad.HsCMV (data not shown).
TABLE 1.
Fluctuation analysis to determine the rate of RCA formation during growth of adenovirus vectors with empty expression cassettes
| Plate no. | Result (FFU/ml [107]) with set indicated for:a
|
|||
|---|---|---|---|---|
| Ad-Con
|
Ad.MmCMV
|
|||
| 1 | 2 | 1 | 2 | |
| 1 | 18.0 | 3.7 | 20 | 40 |
| 2 | 7.6 | 2.9 | 59 | 120 |
| 3 | 1.9 | 3.8 | 21 | 70 |
| 4 | 0.7 | 2.1 | 25 | 53 |
| 5 | 1.1 | 2.0 | 23 | 62 |
| 6 | 1.0 | 1.4 | 32 | 95 |
| 7 | 0.7 | 1.8 | 32 | 69 |
| 8 | 2.2 | 2.7 | 23 | 67 |
| 9 | 6.0 | 3.5 | 34 | 76 |
| 10 | 1.3 | 3.0 | 32 | 74 |
| 11 | 1.9 | 2.8 | 17 | 67 |
| 12 | 1.3 | 2.5 | 22 | 61 |
| Avg | 3.6 | 2.7 | 28 | 71 |
| SD | 5.0 | 0.8 | 11 | 20 |
| Sum | 43.7 | 32.2 | 340 | 854 |
Boldface and italicized titers represent plates containing RCA.
TABLE 4.
RCA ratesa
| Vector | RCA Rate
|
Fold decrease vs Ad-Con | ||
|---|---|---|---|---|
| Set 1 | Set 2 | Avg | ||
| Ad.MmCMV | 1.0 × 10−9 | 1.2 × 10−10 | 5.6 × 10−10 | 45.5 |
| Ad-Con | 2.4 × 10−8 | 2.6 × 10−8 | 2.5 × 10−8 | 1.0 |
| Ad.E2F1 | 1.6 × 10−9 | <1.6 × 10−9b | 1.6 × 10−9 | 3.8 |
| Ad.E2F2 | 3.8 × 10−9 | ND | 3.9 × 10−9 | 6.7 |
| Ad.E2F3 | 3.5 × 10−9 | ND | 3.5 × 10−9 | 7.1 |
| Ad.HsRad51.nls | 2.6 × 10−10 | 4.6 × 10−10 | 3.6 × 10−10 | 71 |
| Ad.SUMO-1 (R to L) | 4.7 × 10−11 | ND | 4.7 × 10−11 | 526 |
| Ad.SUMO-1 (L to R) | <6.7 × 10−11b | ND | <6.7 × 10−11b | >526 |
ND, not done.
Estimate based on the absence of RCA in the 12 or 13 independent stocks of the fluctuation analysis.
It is also clear that the RCA formation rates determined by fluctuation assays are lower than the frequencies of RCA formation. This likely reflects the skew inherent to frequency estimates arising from differential viral growth rates in a mixed RCA/adenovirus vector population and different times of RCA appearance during individual stock production. One-step growth curves were generated for a subset of the vectors tested for fluctuation. Many of these displayed defective growth relative to their empty vector controls (data not shown). Moreover, in the case of RCA generated from Ad.E2F1, Ad.E2F2, and Ad.E2F3, the plaque-initiated RCA stocks replicated better than the vector from which they were derived (data not shown). The differential growth rate of RCA appears to be partly responsible for the observed differences between frequency and rate measurements.
Although the overall viral:293 cell sequence identity is greatest for Ad.HsCMV-based vectors, it is possible that length of sequence identity on one side of a transgene cassette has a greater influence on RCA formation than that of the other side. Although Ad-Con displayed higher RCA formation rates, its adenovirus DNA sequences, located to the right side of the transgene cassette, had a relatively intermediate amount of DNA sequence identity with the Ad5 DNA of 293 cells. On the left side of the transgene cassette, Ad-Con had the least amount of sequence identity (194 bp) with the integrated Ad5 DNA of the 293 cell genome compared to Ad.HsCMV (355 bp) or Ad.MmCMV (456 bp). Because the rates of RCA formation were not based simply on the lengths of identical adenovirus DNA sequences, some other feature(s) had to be causing the increased rate of RCA formation in Ad-Con infected cells.
RCA rates are decreased by the presence or overexpression of E2F1, E2F2, or E2F3 DNA.
RCA generation by 293 cells can also be employed as a tool to define proteins that modulate gene targeting. E2F proteins regulate the expression of a number of genes involved in DNA replication and repair during the transition of the cell cycle from late G1 to S phase. Using microarray-based analysis of Ad.E2F1-infected cells lacking E1A, E2F1 has been shown to induce the transcription of Rad51 (17). Rad51 is the essential central component of the homologous DNA recombination/repair machinery in human cells. Slight increases in Rad51 levels increase HR (45). It was possible that overexpression of E2F1 could increase the rate of cellular recombination and subsequent RCA formation rate. On the other hand, E2F1 expression, induced using a virus-free cell-based system, resulted in a decrease in the microarray-defined transcription of other genes known to interact with Rad51 in the execution of HR and DNA repair (52). These down-regulated recombination proteins included rec2/Rad51B, Rad51C, and Rad52. The frequencies of RCA in the plaque-purified virus stocks of the Ad.E2F family and Ad.DP1 vectors were relatively high. It was possible that high levels of one of the E2F proteins up- or down-regulated genes that altered the rate of RCA recombination. Using a fluctuation test (Table 2), we found that the RCA formation rates of Ad.E2F1, Ad.E2F2, and Ad.E2F3 were similar (Table 4). These rates were lower than that of their empty vector control, Ad-Con. The four- to sevenfold decrease may simply reflect a structural feature of the E2F vector genomes that inhibits recombination. Alternatively, the decreased rate of RCA formation might suggest that expression of any of these E2F proteins negatively affects RCA recombination mechanisms.
TABLE 2.
Fluctuation analysis to determine the rates of RCA formation during growth of Ad.E2F1, Ad.E2F2, and Ad.E2F3
| Plate no. | Result (FFU/ml [107]) for:a
|
|||
|---|---|---|---|---|
| Ad.E2F1
|
Ad.E2F2 | Ad.E2F3 | ||
| Set 1 | Set 2 | |||
| 1 | 14.1 | 9.0 | <1 | 9 |
| 2 | 4.4 | 1.7 | 2 | 3 |
| 3 | 5.8 | 1.7 | <1 | 24 |
| 4 | 5.4 | 1.9 | 14 | 7 |
| 5 | 6.4 | 1.7 | 10 | 9 |
| 6 | 6.8 | 4.1 | 6 | 5 |
| 7 | 5.7 | 11.7 | 1 | 14 |
| 8 | 2.5 | 4.2 | 7 | 6 |
| 9 | 2.2 | 5.3 | 25 | 1 |
| 10 | 2.0 | 7.2 | 67 | 4 |
| 11 | 1.7 | 7.7 | 10 | 6 |
| 12 | 8.9 | 7.5 | 2 | 9 |
| Avg | 5.5 | 5.3 | 14 | 8 |
| SD | 3.5 | 3.3 | 20 | 6 |
| Sum | 65.8 | 63.6 | 142 | 98 |
Boldface and italicized titers represent plates containing RCA.
RCA formation rates are not increased by high-level expression of nuclear-localized HsRad51.
We have been using adenoviruses as tools to study DNA recombination and repair in mammalian cells. Rad51 promotes homologous DNA recombination/repair in association with other cellular proteins, including the Rad51 paralogs (Rad51B, Rad51C, Rad51D, XRCC2, and XRCC3), Rad52, and Rad54 (50). To aid in the investigation of recombination and repair processes, we have constructed adenovirus vectors overexpressing wild-type and mutant forms of these human DNA recombination proteins. The biological properties of adenovirus vectors overexpressing these genes will be reported elsewhere.
It was possible that overproduction of a single protein involved in the multicomponent recombination/repair machine would either increase or decrease RCA formation rate. Table 3 lists RCA fluctuation test results in stocks of Ad.HsRad51.nls, and the rates of RCA formation are listed in Table 4. Ad.HsRad51.nls produces a complete human Rad51 protein with a c-terminal myc tag and three SV40-derived nuclear localization signals. Rad51 lacks a nuclear localization signal, and the addition of the nuclear localization signal assures its nuclear accumulation. The RCA rate during growth of Ad.HsRad51.nls was less than twofold lower than its matched control adenovirus vector (Ad.MmCMV). Furthermore, stocks of adenovirus vectors expressing high levels of other human recombination proteins did not have high levels of RCA. RCA was undetectable in passage 2 plaque-initiated stocks of Ad.HsRad51, Ad.HsRad51B, Ad.HsRad51C, Ad.HsRad51D, Ad.HsXRCC2, Ad.HsXRCC3, Ad.HsRad52, and Ad.HsRad54 when a minimum of 109 FFU per stock was assayed by three sequential passages on A549 cells (data not shown). Fluctuation tests to determine RCA rates were not performed on these vectors, and it is possible that their transgenes may influence RCA formation, but not to the degree of Ad-Con.
TABLE 3.
Fluctuation analysis to determine the rate of RCA formation during growth of Ad.HsRad51.nls
| Plate no. | Result (FFU/ml [107]) for Ad.HsRad51.nls set no.a indicated
|
|
|---|---|---|
| 1 | 2 | |
| 1 | 42 | 110 |
| 2 | 35 | 82 |
| 3 | 49 | 48 |
| 4 | 23 | 38 |
| 5 | 28 | 56 |
| 6 | 29 | 28 |
| 7 | 29 | 59 |
| 8 | 32 | 66 |
| 9 | 31 | 71 |
| 10 | 38 | 85 |
| 11 | 31 | 61 |
| 12 | 25 | 49 |
| Avg | 33 | 63 |
| SD | 7 | 22 |
| Sum | 392 | 753 |
Boldface and italicized titers represent plates containing RCA.
RCA rate is not affected by transcription cassette orientation in vectors with an Ad.MmCMV-like genome structure.
One of the major differences in the adenovirus genomes depicted in Fig. 1B is the direction of transcription from the CMV promoter. The CMV promoters of Ad.HsCMV and Ad.MmCMV direct transcription rightward while Ad-Con cassette transcription is in the leftward direction. It may be that RCA rate is higher in leftward-transcribing cassettes because of interactions between the viral DNA replication, recombination, and transcription machineries located at the left end of the adenovirus genome. To determine if transcription orientation affects RCA formation rate during stock generation, we evaluated stocks of viruses containing a small transgene transcribed from an SV40 expression cassette in either a rightward or a leftward direction. These E1 replacement adenovirus vectors produced similar levels (data not shown) of a His-tagged small ubiquitin-like modifier (SUMO-1) protein, which is involved in posttranslational modification (18). Thirteen stocks were generated for each virus. Five random stocks of each virus construct were titrated to determine the average viral yield per plate. Only one stock of the 26 contained detectable RCA. The transcription orientation of the RCA-positive stock was in the rightward orientation. PCR analysis of DNA from this RCA stock revealed that it contained E1 DNA (data not shown). Moreover, the RCA formation rates of these two vectors were over 500-fold lower than those of Ad-Con (Table 4). The adenovirus portions of these Ad.HisSUMO-1 vectors were identical to those of Ad.MmCMV (Fig. 2), yet Ad.HisSUMO-1 displayed 10-fold-lower RCA formation rates. This difference could be due to the expression of SUMO-1, but it could also be due to the presence of an SV40 expression cassette containing SUMO-1 DNA with different DNA topology near potential sites of recombination. Although we did not invert the orientation of transcription in the Ad-Con vector and measure its effect on RCA generation, the RCA rates of the SUMO-1 vectors support the idea that transcription orientation is not the cause of the relatively high RCA formation rate observed when producing vectors based on Ad-Con.
DISCUSSION
To measure the RCA formation rate of E1 region replacement vectors in 293 cells, we adapted the Luria-Delbrück fluctuation test. A previous report of rate measurement was based on detection of RCA in sequential passages of adenovirus vectors in 293 cells (14).
When choosing a system to measure the rate of RCA formation, there are distinct advantages to analyzing parallel stocks rather than serially passaged stocks. In serial passages, the time of RCA occurrence is subject to chance variations. Also, the rate of RCA replication, compared to that of the parental adenovirus vector, greatly influences the outcome. In parallel stock analyses, the skew caused by time of appearance and differential RCA replication is avoided because a positive or negative outcome is scored simply as the presence or absence of RCA. In the parallel stock fluctuation test for RCA described in this paper, the rate can be determined if enough parallel stocks are tested and if a proportion of the stocks are negative for RCA. Furthermore, the entire viral yield, minus a small amount used for titration on permissive 293 cells, is tested on nonpermissive cells. A549 cells were selected as the nonpermissive cell line and proved to be sensitive indicators of the presence of RCA in a vector stock. We have been able to reproducibly detect a single “spiked” RCA in a plate of A549 cells multiply infected with Ad.HsCMV using a multiplicity of infection of 10 (data not shown).
We tested several variables that might be predicted to increase or decrease RCA rates. The first determined if the overall sequence identity of an adenovirus vector genome with the integrated Ad5 DNA of 293 cells affected the rate of RCA generation. The second determined if the distribution of sequence identity flanking an expression cassette affected RCA rate. The third addressed whether adenovirus vectors, producing high levels of human proteins involved in coordinate regulation of gene expression during transition into the S phase of the cell cycle, exhibited altered recombination rates. The fourth tested if overexpression of Rad51 modified the RCA formation rate. The fifth determined if the direction of transgene cassette transcription altered RCA rate.
Freely replicating Ad5 DNA can acquire Ad2 DNA integrated in the cellular genome of transformed rat F4 cells (6). The events could be easily detected because of DNA sequence and restriction endonuclease site differences between the highly homologous Ad2 and Ad5 genomes. Using an Ad2-based vector, Hehir et al. showed that RCA formation in Ad5-transformed 293 cells also occurred by homologous recombination between the integrated and freely replicating adenovirus DNA (14). Based on DNA sequence variations between Ad2 and Ad5, they reported evidence supporting double crossover events during RCA formation in 293 cells. As would be predicted by HR mechanisms, the exchanges were located in regions of adenovirus DNA flanking the expression cassettes of the input vector genomes. They also reported that the amount of homology flanking an individual transgene cassette correlated with the amount of RCA detected.
Based on overall sequence identity, the RCA rate would be predicted to be higher in stocks of Ad.HsCMV and lower in Ad.MmCMV or Ad-Con, which have sequence identities with the 293 genome of 1,733, 1,266 and 1,210 bp, respectively (Fig. 1B and Table 5). In contrast to the prediction, our measurements of RCA rates and frequencies suggested that overall sequence identity was not the basis of the relatively high RCA rate in Ad-Con stocks compared to Ad.MmCMV and Ad.HsCMV. Also, it can be argued that recombination frequencies will differ on each side of the transgene cassette based on lengths of sequence identity. Because RCA recombination requires a double crossover event, the recombination frequency should be more limited by the side of the expression cassette with the smaller length of homology. Again, in contrast to expectation, Ad-Con vectors, which would be predicted to have the lowest rate of recombination, had the highest rate of RCA formation.
TABLE 5.
Relationships of RCA rates predicted from overall and distributed sequence identities to observed RCA formation rates
| Vector | Identity (bp)
|
Prediction
|
Observed RCA rate | |||
|---|---|---|---|---|---|---|
| Left | Right | Overall | Overall | Distributed | ||
| Ad.HsCMV | 355 | 1,378 | 1,733 | Higher | Intermediate | Lower |
| Ad.MmCMV | 456 | 810 | 1,266 | Lower | Highest | Lower |
| Ad-Con | 194 | 1,016 | 1,210 | Lower | Lowest | Higher |
In addition to sequence identity, the vectors differ in the location of the packaging signal used to assemble DNA into virions. In the Ad-Con-derived vectors, the packaging signal has been moved to the right end of the genome, while it is retained in its natural location in the Ad.MmCMV and Ad.HsCMV vectors. Although we have not tested if the packaging signal can interfere with recombination and RCA formation, it is possible that viral proteins interact with the packaging signal and interfere with recombination.
Infecting cells with adenovirus vectors overexpressing transcription factors that increase or decrease the expression of DNA recombination genes did not increase the rate of RCA formation. We found that infection of 293 cells by adenovirus vectors expressing E2F1, E2F2, or E2F3 resulted in a four- to sevenfold decrease in the rate of RCA formation compared to Ad-Con, the empty vector control. Several mechanisms that could explain this decrease include the following: (i) either positive or negative regulation, by E2F transcription factors, of genes involved in controlling HR; (ii) E2F responses, such as apoptosis, positive or negative regulation of cell cycle progression, differentiation, induction of checkpoints, DNA synthesis, and DNA replication (reviewed in reference 3); (iii) “stuffer” DNA causing changes in local DNA conformation and/or increasing the distance between the two crossover events; and (iv) longer transcription runs from expression cassettes containing a transgene.
To study recombination proteins in vivo, vectors overexpressing HsRad51, HsRad51B, HsRad51C, HsRad51D, HsXRCC2, HsXRCC3, HsRad52, and HsRad54 were constructed. Preliminary RCA screening of these stocks has not detected RCA to the level of Ad-Con (data not shown). Small increases in Rad51 protein levels cause an increase in DNA recombination (45). In contrast, the infection of 293 cells with Ad.HsRad51.nls resulted in a slight decrease in the RCA rate (1.6-fold) compared to that of the matching empty vector control, Ad.MmCMV.
An obvious difference in the vectors, which could explain the observed differences in RCA formation rates, is the direction of expression cassette transcription. It was possible that transcription aimed toward the left end of the adenovirus genome would open up this region to recombination as originally proposed for other systems (1, 41, 46; reviewed in reference 5). We constructed adenovirus vectors expressing His-tagged SUMO-1 transcribed in rightward or leftward directions and tested these in a fluctuation test. Ad.HisSUMO-1 vectors contained an SV40 enhancer-driven expression cassette with flanking adenovirus DNA identical to that of Ad.MmCMV. Although we did not reverse the transcription cassette of Ad-Con to determine if leftward transcription caused the relative increase in RCA rate in this family of vectors, the RCA formation rate of the leftward-transcribed Ad.HisSUMO-1 vector was not higher than that of the vector expressing HisSUMO-1 in the rightward direction, suggesting transcription direction does not influence RCA generation.
RCA should be absent or kept to a minimum in adenovirus vector stocks intended for clinical applications. Better RCA rate measurements allowed meaningful comparisons of RCA formation in different families of E1-substituted vectors in the presence or absence of highly expressed transgenes. These findings underscore the point that one cannot predict RCA based simply on the extent of sequence identity with the integrated DNA of 293 cells. This caution should be considered when designing new vectors that will be produced in 293 cells. Performing the RCA fluctuation test on 293 cell-produced adenovirus vectors destined for patients would provide indicators of RCA susceptibility and suitability for pharmaceutical use.
Although 293 cells are widely used for adenovirus vector construction and propagation, other cell lines have been constructed that express E1A and E1B but avoid any or extensive sequence homologies (11, 16, 40). These cells do not provide homologous targets for recombination. Unusual nonhomologous recombination events have been reported for PER.C6 cells that result in a small replicating adenovirus genome that can replicate and package within nonpermissive cells only in the presence of the original adenovirus vector (31). These are not typical RCA in that they cannot be plaque purified and cannot replicate efficiently at a low multiplicity of infection in A549 cells. It is clear that although 293 cells are useful for the production of today's vector preparations, better cell lines or vectors will be used to diminish RCA.
RCA formation is a targeted recombination event. Adenovirus vectors represent easily manipulated tools that can be used to study the rate of recombination in 293 cells. They can be used to simultaneously overexpress wild-type or altered recombination proteins. They can also be used to overexpress genes, such as transcription factors, suspected of regulating recombination gene expression. Defining the factors that regulate RCA formation in 293 cells will improve the understanding of molecular mechanisms governing recombination and should contribute to the development of human gene targeting systems.
Acknowledgments
We thank Joseph Nevins for Ad.E2F1, Ad.E2F2, Ad.E2F3, Ad.E2F4, Ad.E2F5, and Ad.DP1; Efim Golub for HsRad51 cDNA; Richard Fishel for HsRad54 cDNA; Andrew Yueh for pSG5-HisSUMO-1; and Patrick Sung for HsRad52, HsRad51C, HsXRCC2, and HsXRCC3 cDNAs.
This work was supported in part by grant GM31452 to C.S.H.Y. from the NIH and by grant CA13696 awarded to Columbia Comprehensive Cancer Center.
REFERENCES
- 1.Blackwell, T. K., M. W. Moore, G. D. Yancopoulos, H. Suh, S. Lutzker, E. Selsing, and F. W. Alt. 1986. Recombination between immunoglobulin variable region gene segments is enhanced by transcription. Nature 324:585-589. [DOI] [PubMed] [Google Scholar]
- 2.Boyer, J., K. Rohleder, and G. Ketner. 1999. Adenovirus E4 34k and E4 11k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology 263:307-312. [DOI] [PubMed] [Google Scholar]
- 3.Bracken, A. P., M. Ciro, A. Cocito, and K. Helin. 2004. E2F target genes: unraveling the biology. Trends Biochem. Sci. 29:409-417. [DOI] [PubMed] [Google Scholar]
- 4.Carson, C. T., R. A. Schwartz, T. H. Stracker, C. E. Lilley, D. V. Lee, and M. D. Weitzman. 2003. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 22:6610-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaudhuri, J., and F. W. Alt. 2004. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat. Rev. Immunol. 4:541-552. [DOI] [PubMed] [Google Scholar]
- 6.Duigou, G. J., and S. G. Zimmer. 1983. Use of a viral probe to study recombinational exchanges in mammalian cells. Prog. Nucleic Acid Res. Mol. Biol. 29:137-140. [DOI] [PubMed] [Google Scholar]
- 7.Dunn, A. R., P. H. Gallimore, K. W. Jones, and J. K. McDougall. 1973. In situ hybridization of adenovirus RNA and DNA. II. Detection of adenovirus-specific DNA in transformed and tumour cells. Int. J. Cancer 11:628-636. [DOI] [PubMed] [Google Scholar]
- 8.Endter, C., and T. Dobner. 2004. Cell transformation by human adenoviruses. Curr. Top. Microbiol. Immunol. 273:163-214. [DOI] [PubMed] [Google Scholar]
- 9.Ensinger, M. J., and H. S. Ginsberg. 1972. Selection and preliminary characterization of temperature-sensitive mutants of type 5 adenovirus. J. Virol. 10:328-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans, J. D., and P. Hearing. 2003. Distinct roles of the Adenovirus E4 ORF3 protein in viral DNA replication and inhibition of genome concatenation. J. Virol. 77:5295-5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fallaux, F. J., A. Bout, I. van der Velde, D. J. van den Wollenberg, K. M. Hehir, J. Keegan, C. Auger, S. J. Cramer, H. van Ormondt, A. J. van der Eb, D. Valerio, and R. C. Hoeben. 1998. New helper cells and matched early region 1-deleted adenovirus vectors prevent generation of replication-competent adenoviruses. Hum. Gene Ther. 9:1909-1917. [DOI] [PubMed] [Google Scholar]
- 12.Flint, S. J., S. M. Berget, and P. A. Sharp. 1976. Characterization of single-stranded viral DNA sequences present during replication of adenovirus types 2 and 5. Cell 9:559-571. [DOI] [PubMed] [Google Scholar]
- 13.Harui, A., S. Suzuki, S. Kochanek, and K. Mitani. 1999. Frequency and stability of chromosomal integration of adenovirus vectors. J. Virol. 73:6141-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hehir, K. M., D. Armentano, L. M. Cardoza, T. L. Choquette, P. B. Berthelette, G. A. White, L. A. Couture, M. B. Everton, J. Keegan, J. M. Martin, D. A. Pratt, M. P. Smith, A. E. Smith, and S. C. Wadsworth. 1996. Molecular characterization of replication-competent variants of adenovirus vectors and genome modifications to prevent their occurrence. J. Virol. 70:8459-8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 16.Imler, J. L., C. Chartier, D. Dreyer, A. Dieterle, M. Sainte-Marie, T. Faure, A. Pavirani, and M. Mehtali. 1996. Novel complementation cell lines derived from human lung carcinoma A549 cells support the growth of E1-deleted adenovirus vectors. Gene Ther. 3:75-84. [PubMed] [Google Scholar]
- 17.Ishida, S., E. Huang, H. Zuzan, R. Spang, G. Leone, M. West, and J. R. Nevins. 2001. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 21:4684-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, E. S. 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73:355-382. [DOI] [PubMed] [Google Scholar]
- 19.Kelly, T. J. 1984. Adenovirus DNA replication, p. 271-308. In H. S. Ginsberg (ed.), The adenoviruses. Plenum Press, New York, N.Y.
- 20.Kovesdi, I., R. Reichel, and J. R. Nevins. 1987. Role of an adenovirus E2 promoter binding factor in E1A-mediated coordinate gene control. Proc. Natl. Acad. Sci. USA 84:2180-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawrence, W. C., and H. S. Ginsberg. 1967. Intracellular uncoating of type 5 adenovirus deoxyribonucleic acid. J. Virol. 1:851-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lochmuller, H., A. Jani, J. Huard, S. Prescott, M. Simoneau, B. Massie, G. Karpati, and G. Acsadi. 1994. Emergence of early region 1-containing replication-competent adenovirus in stocks of replication-defective adenovirus recombinants (delta E1 + delta E3) during multiple passages in 293 cells. Hum. Gene Ther. 5:1485-1491. [DOI] [PubMed] [Google Scholar]
- 23.Louis, N., C. Evelegh, and F. L. Graham. 1997. Cloning and sequencing of the cellular-viral junctions from the human adenovirus type 5 transformed 293 cell line. Virology 233:423-429. [DOI] [PubMed] [Google Scholar]
- 24.Luria, S., and M. Delbrück. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma, Y., R. Croxton, R. L. Moorer, Jr., and W. D. Cress. 2002. Identification of novel E2F1-regulated genes by microarray. Arch. Biochem. Biophys. 399:212-224. [DOI] [PubMed] [Google Scholar]
- 26.McDougall, J. K., P. H. Gallimore, A. R. Dunn, and K. W. Jones. 1972. Adenoviruses in tumour cells. Lancet 1:1022-1023. [DOI] [PubMed] [Google Scholar]
- 27.McGrory, W. J., D. S. Bautista, and F. L. Graham. 1988. A simple technique for the rescue of early region I mutations into infectious human adenovirus type 5. Virology 163:614-617. [DOI] [PubMed] [Google Scholar]
- 28.Meinschad, C., and E. L. Winnacker. 1980. Recombination in adenovirus. I. Analysis of recombinant viruses under non-selective conditions. J. Gen. Virol. 48:219-224. [DOI] [PubMed] [Google Scholar]
- 29.Mitani, K., and S. Kubo. 2002. Adenovirus as an integrating vector. Curr. Gene Ther. 2:135-144. [DOI] [PubMed] [Google Scholar]
- 30.Mohammadi, E. S., E. A. Ketner, D. C. Johns, and G. Ketner. 2004. Expression of the adenovirus E4 34k oncoprotein inhibits repair of double strand breaks in the cellular genome of a 293-based inducible cell line. Nucleic Acids Res. 32:2652-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murakami, P., M. Havenga, F. Fawaz, R. Vogels, G. Marzio, E. Pungor, J. Files, L. Do, J. Goudsmit, and M. McCaman. 2004. Common structure of rare replication-deficient E1-positive particles in adenoviral vector batches. J. Virol. 78:6200-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng, P., D. T. Cummings, C. M. Evelegh, and F. L. Graham. 2000. Yeast recombinase FLP functions effectively in human cells for construction of adenovirus vectors. BioTechniques 29:524-526, 528. [DOI] [PubMed] [Google Scholar]
- 33.Nicolás, A. L., P. L. Munz, E. Falck-Pedersen, and C. S. H. Young. 2000. Creation and repair of specific DNA double-strand breaks in vivo following infection with adenovirus vectors expressing Saccharomyces cerevisiae HO endonuclease. Virology 266:211-224. [DOI] [PubMed] [Google Scholar]
- 34.Philipson, L. 1961. Adenovirus assay by the fluorescent cell-counting procedure. Virology 15:263-268. [DOI] [PubMed] [Google Scholar]
- 35.Polager, S., Y. Kalma, E. Berkovich, and D. Ginsberg. 2002. E2Fs up-regulate expression of genes involved in DNA replication, DNA repair and mitosis. Oncogene 21:437-446. [DOI] [PubMed] [Google Scholar]
- 36.Ren, B., H. Cam, Y. Takahashi, T. Volkert, J. Terragni, R. A. Young, and B. D. Dynlacht. 2002. E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes Dev. 16:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rouet, P., F. Smih, and M. Jasin. 1994. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol. 14:8096-8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., M. Sleigh, J. A. Engler, and T. R. Broker. 1980. The evolution of the adenoviral genome. Ann. N. Y. Acad. Sci. 354:426-452. [DOI] [PubMed] [Google Scholar]
- 39.Sancar, A., L. A. Lindsey-Boltz, K. Unsal-Kaccmaz, and S. Linn. 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73:39-85. [DOI] [PubMed] [Google Scholar]
- 40.Schiedner, G., S. Hertel, and S. Kochanek. 2000. Efficient transformation of primary human amniocytes by E1 functions of Ad5: generation of new cell lines for adenoviral vector production. Hum. Gene Ther. 11:2105-2116. [DOI] [PubMed] [Google Scholar]
- 41.Serizawa, N., T. Horiuchi, and T. Kobayashi. 2004. Transcription-mediated hyper-recombination in HOT1. Genes Cells 9:305-315. [DOI] [PubMed] [Google Scholar]
- 42.Smith, D. S., G. Leone, J. DeGregori, M. N. Ahmed, M. B. Qumsiyeh, and J. R. Nevins. 2000. Induction of DNA replication in adult rat neurons by deregulation of the retinoblastoma/E2F G1 cell cycle pathway. Cell Growth Differ. 11:625-633. [PubMed] [Google Scholar]
- 43.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348-352. [DOI] [PubMed] [Google Scholar]
- 44.Thiry, M., and F. Puvion-Dutilleul. 1995. Differential distribution of single-stranded DNA, double-stranded DNA, and RNA in adenovirus-induced intranuclear regions of HeLa cells. J. Histochem. Cytochem. 43:749-759. [DOI] [PubMed] [Google Scholar]
- 45.Vispé, S., C. Cazaux, C. Lesca, and M. Defais. 1998. Overexpression of Rad51 protein stimulates homologous recombination and increases resistance of mammalian cells to ionizing radiation. Nucleic Acids Res. 26:2859-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voelkel-Meiman, K., R. L. Keil, and G. S. Roeder. 1987. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell 48:1071-1079. [DOI] [PubMed] [Google Scholar]
- 47.Volkert, F. C., and C. S. H. Young. 1983. The genetic analysis of recombination using adenovirus overlapping terminal DNA fragments. Virology 125:175-193. [DOI] [PubMed] [Google Scholar]
- 48.Weiden, M. D., and H. S. Ginsberg. 1994. Deletion of the E4 region of the genome produces adenovirus DNA concatemers. Proc. Natl. Acad. Sci. USA 91:153-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weitzman, M. D., C. T. Carson, R. A. Schwartz, and C. E. Lilley. 2004. Interactions of viruses with the cellular DNA repair machinery. DNA Repair 3:1165-1173. [DOI] [PubMed] [Google Scholar]
- 50.West, S. C. 2003. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 4:435-445. [DOI] [PubMed] [Google Scholar]
- 51.Williams, J. F., and S. Ustaçelebi. 1971. Complementation and recombination with temperature-sensitive mutants of adenovirus type 5. J. Gen. Virol. 13:345-348. [DOI] [PubMed] [Google Scholar]
- 52.Young, A. P., R. Nagarajan, and G. D. Longmore. 2003. Mechanisms of transcriptional regulation by Rb-E2F segregate by biological pathway. Oncogene 22:7209-7217. [DOI] [PubMed] [Google Scholar]
- 53.Young, C. S. H. 1995. Homologous recombination in the replicative cycle of adenovirus and its relationship to DNA replication. Curr. Top. Microbiol. Immunol. 199(Pt. 2):89-108. [DOI] [PubMed] [Google Scholar]
- 54.Young, C. S. H., and G. J. Duigou. 1999. Homologous recombination between exogenous and integrated adenovirus DNA sequences, p. 229-236. In P. Seth (ed.), Adenoviruses: basic biology to gene therapy. R. G. Landes Co., Austin, Tex.
- 55.Young, C. S. H., and P. B. Fisher. 1980. Adenovirus recombination in normal and repair-deficient human fibroblasts. Virology 100:179-184. [DOI] [PubMed] [Google Scholar]
- 56.Young, C. S. H., A. L. Nicolás, H. Lu, and P. L. Munz. 1999. Methods for creating and analyzing adenovirus vectors that express proteins that act on the viral genome, p. 61-84. In W. S. M. Wold (ed.), Adenovirus methods and protocols. Humana Press, Totowa, N.J.
- 57.Young, C. S. H., and S. J. Silverstein. 1980. The kinetics of adenovirus recombination in homotypic and heterotypic genetic crosses. Virology 101:503-515. [DOI] [PubMed] [Google Scholar]