Abstract
To succeed, many cells must alternate between life-styles that permit rapid growth in the presence of abundant nutrients and ones that enhance survival in the absence of those nutrients. One such change in life-style, the “acetate switch,” occurs as cells deplete their environment of acetate-producing carbon sources and begin to rely on their ability to scavenge for acetate. This review explains why, when, and how cells excrete or dissimilate acetate. The central components of the “switch” (phosphotransacetylase [PTA], acetate kinase [ACK], and AMP-forming acetyl coenzyme A synthetase [AMP-ACS]) and the behavior of cells that lack these components are introduced. Acetyl phosphate (acetyl∼P), the high-energy intermediate of acetate dissimilation, is discussed, and conditions that influence its intracellular concentration are described. Evidence is provided that acetyl∼P influences cellular processes from organelle biogenesis to cell cycle regulation and from biofilm development to pathogenesis. The merits of each mechanism proposed to explain the interaction of acetyl∼P with two-component signal transduction pathways are addressed. A short list of enzymes that generate acetyl∼P by PTA-ACKA-independent mechanisms is introduced and discussed briefly. Attention is then directed to the mechanisms used by cells to “flip the switch,” the induction and activation of the acetate-scavenging AMP-ACS. First, evidence is presented that nucleoid proteins orchestrate a progression of distinct nucleoprotein complexes to ensure proper transcription of its gene. Next, the way in which cells regulate AMP-ACS activity through reversible acetylation is described. Finally, the “acetate switch” as it exists in selected eubacteria, archaea, and eukaryotes, including humans, is described.
INTRODUCTION
Definitions
To survive, many cells must switch from a physiological program that permits rapid growth in the presence of abundant nutrients to one that enhances survival in the absence of those nutrients. One such “switch” occurs when bacterial cells transit from a program of rapid growth that produces and excretes acetate (dissimilation) to a program of slower growth facilitated by the import and utilization (assimilation) of that excreted acetate (Fig. 1). This “acetate switch” occurs as cells deplete their environment of acetate-producing (acetogenic) carbon sources, e.g., d-glucose or l-serine, and begin to rely on their ability to scavenge for environmental acetate.
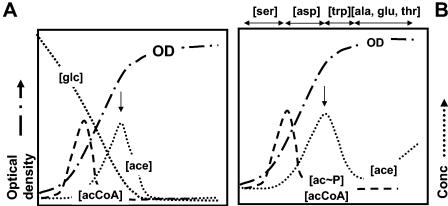
FIG. 1.
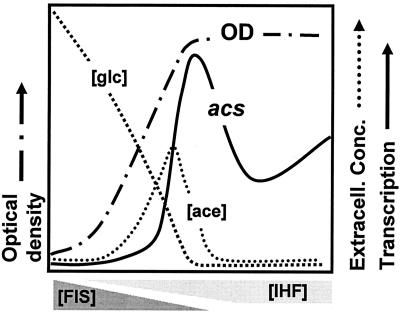
Schematics showing the “acetate switch” during aerobic growth in minimal medium supplemented with glucose as the sole carbon source (A) and in tryptone broth (B). The single-headed arrow points to the physiological acetate switch. OD, optical density. [glc] and [ace], extracellular glucose and acetate concentrations. [ser], [asp], [trp], [ala], [glu], and [thr], extracellular amino acid concentrations. The double-headed arrows denote the interval of amino acid consumption. [acCoA] and [ac∼P], intracellular acetyl-CoA and acetyl∼P concentrations.
Throughout this review, I define the “acetate switch” physiologically as the moment when acetate dissimilation equals its assimilation. One observes this event experimentally as the peak accumulation of extracellular acetate (indicated by single-headed arrows in Fig. 1). Note that this physiological event cannot occur unless a molecular “switch” already has been “flipped” to express and activate the machinery responsible for acetate assimilation.
The Physiological “Switch”: Three Examples
The “acetate switch” of Escherichia coli has been studied predominantly under three different growth conditions: in shake flask culture supplemented with d-glucose as the sole carbon source, in shake flask culture with a tryptone-based medium, and during high-cell-density glucose fermentation. The following simply describes the “switch” as it occurs under each regimen and does not attempt to explain the underlying mechanism(s). Such explanations will follow.
Shake flask culture: glucose.
Cells undergo an “acetate switch” during buffered growth on d-glucose (Fig. 1A). During exponential growth, cells consume the sugar and dissimilate acetate (195). Before they exhaust the sugar, however, the “switch” occurs and the cells coassimilate both acetate and the remaining sugar (325). This “switch” occurs just as the cells begin the transition to stationary phase, defined in this review as the moment that the cells begin to decelerate their growth.
Shake flask culture: tryptone.
During exponential growth on Bacto Tryptone broth (an unbuffered, essentially carbohydrate-free mixture of amino acids and small peptides), cells of E. coli consume amino acids in a strictly preferential order (Fig. 1B). First, they consume l-serine and then l-aspartate, while dissimilating acetate, which acidifies the unbuffered medium. As these cells begin the transition to stationary phase, they consume l-tryptophan while assimilating acetate. This consumption of acetate, combined with evolution of ammonia from amino acid metabolism, alkalinizes the medium. On entry into stationary phase, the cells consume a mixture of amino acids, two acetogenic (l-threonine and l-alanine) and one nonacetogenic (l-glutamate). The result is net acetate excretion, albeit to levels lower than those achieved during exponential growth. Because of the continued evolution of ammonia, the environment remains alkaline (78, 358, 359). Thus, the physiological switch can flip back and forth depending on the acetogenic nature of the amino acid(s) presently under consumption.
Glucose-fed high-cell-density fermentation.
The feeding of glucose in a nonlimiting manner to an aerobic fermentation (buffered at pH 7.0) results in an extended growth phase. This extended growth phase results in high cell density accompanied by excretion of large amounts of acetate. These glucose-fed fermentations begin with a glucose-consuming, acetogenic exponential phase during which oxygen is consumed and carbon dioxide evolves. Near the end of exponential growth, the fermentation pauses for a short interval. During this pause, cells halt the consumption of oxygen and the evolution of carbon dioxide. After about 30 min, fermentation reinitiates. Oxygen consumption and carbon dioxide evolution resume as the culture cometabolizes glucose and acetate. Despite buffering, a transient increase in pH accompanies the consumption of acetate (241).
Short History
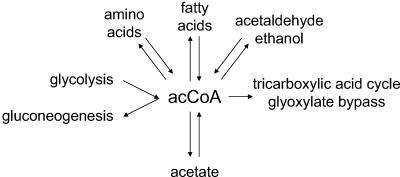
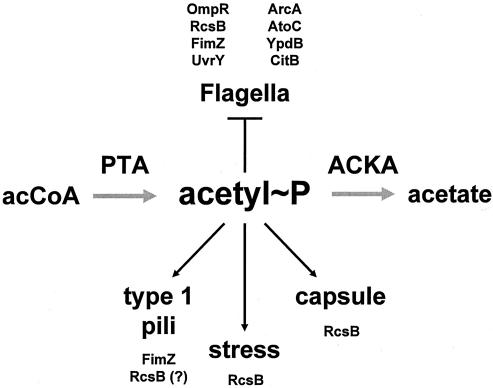
The “acetate switch” possesses a rich past. Its components and intermediates were discovered and initially characterized in the 1940s and 1950s, during the effort to identify the “activated acetate.” We now know this “activated acetate” as acetyl coenzyme A (acetyl-CoA), the high-energy intermediate that sits at the crossroads of central metabolism (Fig. 2) (for historical reviews, see references 32, 45, 231, and 404). During the next three decades, as researchers explored the fundamentals of molecular biology, studies of acetate metabolism faded from prominence, kept alive mostly by investigators concerned with fermentation. On occasion, general interest in the “switch” resurfaced transiently; however, it was not until the late 1980s and early 1990s that the “acetate switch” regained the spotlight. This renewed interest resulted primarily from the proposition that acetyl phosphate (acetyl∼P), the high-energy intermediate of the dissimilation pathway, might function as a global signal (298, 463). Today, mounting evidence suggests that, indeed, acetyl∼P plays such a role, regulating cellular processes as diverse as nitrogen assimilation, osmoregulation, flagellar biogenesis, pilus assembly, capsule biosynthesis, biofilm development, and pathogenicity (24, 25, 187, 273, 274, 291, 322, 343, 354, 355, 358, 473).
FIG. 2.
Acetyl-CoA (acCoA) sits at the crossroads of central metabolism.
For several reasons, there also exists renewed interest in the acetate assimilation enzyme AMP-forming acetyl-CoA synthetase (AMP-ACS). First, AMP-ACS is a prototype for enzymes involved in the synthesis of fatty acids, some antibiotics, and certain anticancer drugs, as well as the degradation of pollutants (427). Second, AMP-ACS activity is regulated by an acetylation-deacetylation system homologous to that used by eukaryotes to control chromatin structure, silencing, mitochondrial signaling, and aging (72, 427). Third, the complex acs promoter that drives AMP-ACS expression in E. coli is fast becoming a model for how dynamic nucleoprotein complexes ensure that transcription occurs properly (33, 40, 62, 63).
Scope
The purpose of this review is to (re)introduce the “acetate switch,” first giving a brief description of the acetate-rich colon, a key ecological niche for E. coli, and then explaining why, when, and how bacterial cells excrete acetate and other central metabolic intermediates. It will acquaint the reader with the enzyme components that comprise the molecular core of the “switch” and describe the behavior of mutants that lack some or all of those components. This review does not, however, summarize the structure-function relationships of these components. Next, emphasis shifts to acetyl∼P, the high-energy intermediate of acetate dissimilation. The review describes how cells regulate the size of the acetyl∼P pool, presents evidence that acetyl∼P can act as a global signal that influences diverse cellular processes, and addresses the mechanism(s) by which acetyl∼P might exert its influence. Next, it focuses on the molecular mechanisms that facilitate the “switch” from acetate dissimilation to acetate assimilation. These mechanisms regulate transcription from the complex acs promoter and the activity of AMP-ACS. Finally, it describes variants of the “acetate switch” found in other eubacterial species, selected archaea, and humans. Although this review does not exhaustively review the literature concerning acid and organic acid stress, the topic is addressed in passing. It also does not directly review efforts to metabolically engineer the “acetate switch,” although much of the information provided will aid researchers interested in such endeavors.
LIFE IN THE COLON
In utero, the mammalian fetus is sterile (303). During and after birth, the human neonate becomes exposed to and colonized by large numbers of E. coli and Enterococcus organisms (108 to 1010 per g of contents). These rapidly growing, facultative bacteria metabolize the lactose present in breast milk to acetate and other short-chain fatty acids (SCFA, also known as volatile fatty acids), creating a reduced acidic environment favorable for colonization by the slower-growing, anaerobic acidophile Bifidobacterium. This acidophilic anaerobe eventually outcompetes E. coli, consumes most of the available sugar, and excretes large amounts of acetate and other SCFA (306).
Subsequent neonatal exposure to microbes through diet, and the competition between these microbes for limited nutrients and specific niches, cause the colonic microbiota to diversify (142, 143). The diversified adult colon hosts a complex flora, consisting of more than 50 genera and 400 species, generally attached as biofilms to particulate intestinal materials and embedded in the mucus layer that coats the colonic epithelial cells (colonocytes). This flora includes large numbers (1010 to 1011 per g) of anaerobes. It also contains a smaller number of facultative organisms, including E. coli, that maintain the reduced environment required by strict anaerobes. Some of these anaerobes, primarily members of the genera Bifidobacterium and Bacteroides, ferment dietary fiber—complex polysaccharides not digested and absorbed in the upper gut—to simple sugars and SCFA. Other anaerobes, mainly Peptostreptococcus and Fusobacterium, as well as certain facultative organisms, e.g., Enterococcus and E. coli, presumably cross-feed on those simple sugars and SCFA (101, 283, 284, 341).
In the colon, acetogenic sugars arise from diet, bacterial metabolism, or the host-secreted mucus (143, 352, 460). This mucus, which overlays colonocytes, is a complex gel of glycolipids, glycoproteins, and a variety of sugar residues, including N-acetylglucosamine, N-acetylgalactosamine, d-galactose, fucose, sialic acids, glucuronate, galacturonate, and gluconate (5). When stripped from mucus, these sugar residues provide carbon that permits rapid growth while contributing to the local accumulation of acetate and other SCFA. As in culture, the nature of the carbon source(s) dictates colonic pH, which can vary from 6 to 8 (14, 55, 102).
SCFA constitute approximately two-thirds of the colonic anion concentration (70 to 130 mM), mainly as acetate, propionate, and butyrate. Of these, acetate predominates. These SCFA, rapidly absorbed by the colonic mucosa, represent the primary energy source for colonocytes, hepatic cells, fat cells, and muscle cells (283, 284, 300, 313, 447). SCFA also perform functions of considerable significance to the health of the host (36, 37, 352, 397, 460). Finally, they enhance the virulence of certain enteric pathogens, including E. coli and other members of the Enterobacteriaceae, mostly by inducing protective mechanisms (39, 122, 255, 260, 270, 377).
WHY CELLS EXCRETE ACETATE
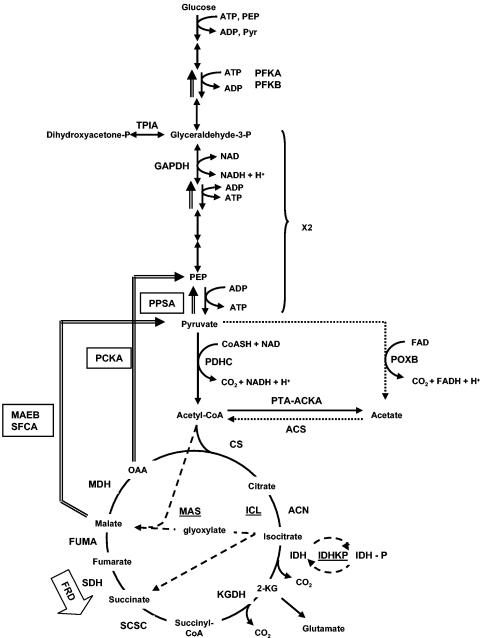
Acetogenesis, the excretion of acetate into the environment, results from the need to regenerate the NAD+ consumed by glycolysis and to recycle the coenzyme A (CoASH) required to convert pyruvate to acetyl-CoA. Since the tricarboxylic acid (TCA) cycle completes the oxidation of acetyl-CoA to carbon dioxide, acetogenesis occurs whenever the full TCA cycle does not operate or when the carbon flux into cells exceeds its capacity and that of other central metabolic pathways (78, 123, 126, 195, 196, 232, 263, 286, 386, 458, 477). Thus, acetate excretion occurs anaerobically during mixed-acid fermentation (54). It also occurs aerobically when growth on excess glucose (or other highly assimilable carbon sources) inhibits respiration (195, 196), a behavior called the bacterial Crabtree effect (98, 119, 280, 379). As a consequence of the Crabtree effect, as much as 15% of the glucose can be excreted as acetate (196). Although acetogenesis has long been considered simply the result of “overflow” metabolism (195, 196), a recent report raises the possibility that acetate excretion permits more rapid growth to higher cell densities primarily by providing the TCA cycle enzyme 2-ketoglutarate dehydrogenase (KGDH) (Fig. 3) with CoASH (124).
FIG. 3.
The pathways of central metabolism. For glycolysis, only some of the intermediates and enzymes of glycolysis are noted. PEP, phosphoenolpyruvate; Pyr, pyruvate; PFK, phosphofructokinase; TPIA, triosephosphate isomerase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PDHC, pyruvate dehydrogenase complex. For acetate metabolism: POXB, pyruvate oxidase; PTA-ACKA, phosphotransacetylase-acetate kinase pathway; ACS, AMP-forming acetyl-CoA synthetase. The dotted arrows denote the proposed PDHC bypass formed by POXB and AMP-ACS. For the TCA cycle: CS, citrate synthase; ACN, aconitase; IDH, isocitrate dehydrogenase; 2-KG, 2-ketoglutarate; KGDH, 2-ketoglutarate dehydrogenase; SCSC, succinyl-CoA synthetase complex; SDH, succinate dehydrogenase; FUMA, fumarase; MDH, malate dehydrogenase; OAA, oxaloacetate. FRD, fumarate reductase, expressed under anaerobic conditions, bypasses SDH. For the glyoxylate bypass: ICL, isocitrate lyase; MAS, malate synthase; IDHK/P, isocitrate dehydrogenase kinase/phosphatase. Underlines and dashed arrows denote enzymes and steps unique to the glyoxylate bypass. For gluconeogenesis: PPSA, PEP synthase; PCKA, pyruvate carboxykinase; MAEB and SFCA, malic enzymes. Boxes and double-lined arrows denote enzymes and steps unique to gluconeogenesis.
Limiting the Tricarboxylic Acid Cycle
The availability of oxygen and the nature and quantity of the carbon source dictate the status of the TCA cycle (Fig. 3) (6, 160, 420). In the absence of oxygen and under conditions that favor catabolite repression (e.g., excess glucose), E. coli cells do not induce the full TCA cycle (6, 320, 342). Instead, they operate a branched version, which forms succinyl-CoA by a reductive pathway and 2-ketoglutarate by an oxidative one (100, 168, 282, 420). This branched form of the TCA cycle does not generate energy; instead it functions biosynthetically, producing precursor metabolites. Thus, ATP must come from glycolysis (6) and substrate phosphorylation via the phosphotransacetylase (PTA)-acetate kinase (ACKA) pathway (61, 385, 441).
This branched version occurs because the absence of oxygen severely inhibits the expression of many TCA cycle enzymes, but most dramatically succinate dehydrogenase (SDH), the succinyl-CoA synthetase complex (SCSC), and KGDH. A more moderate inhibition occurs in the presence of excess glucose (160, 170, 193, 212, 335-339, 413).
In the absence of oxygen, the oxygen-sensitive global regulators ArcA and FNR mediate the repression of many TCA promoters, but most dramatically the sdh-suc operon, which encodes SDH, KDGH, and the SCSC (104, 105, 170, 293, 335, 339, 406). The mechanism that causes glucose repression, and thus the Crabtree effect, is less clear. Glucose represses sdh-suc indirectly through the action of EIICB(Glc), but the mechanism remains a mystery (439). The membrane-bound EIICB(Glc), part of the phosphoenolpyruvate (PEP):carbohydrate phosphotransferase system (PTS), phosphorylates and translocates glucose. This process causes dephosphorylation of EIICB(Glc) (351). Dephosphorylated EIICB(Glc) sequesters Mlc, a global repressor of genes that encode certain sugar-metabolizing enzymes and their uptake systems, including ptsG, the gene that encodes EIICB(Glc). Thus, translocation of glucose by EIICB(Glc) relieves Mlc-dependent repression, which permits expression of these glucose-activated genes. For a review, see reference 349.
For sdh-suc and other glucose-repressed promoters, however, the mechanism remains murky. The process does not involve Mlc directly (439), nor does it involve perturbations in the PEP pool (81, 82). Glucose translocation by EIICB(Glc) dephosphorylates EIIA(Glc), the immediate phosphoryl donor of the PTS. Dephosphorylated EIIA(Glc) causes inducer exclusion, by binding and inhibiting other sugar permeases (for reviews, see references 351 and 434). Dephosphorylation of EIIA(Glc) also may reduce cyclic AMP (cAMP) levels by reducing the activity of adenylate cyclase (351, 434), although this model has been disputed (239). However, despite the presence of putative DNA binding sites for the cAMP receptor protein (CRP) (472), it appears that cAMP-CRP does not control sdh-suc transcription directly (339). Apparently, another global carbon regulator, Cra (also known as FruR), also is not involved (339). Attempts to solve this puzzle are clearly warranted, especially given the negative effect exerted by acetate on the production of recombinant products during aerobic glucose-fed high-cell-density fermentations (15, 38, 174, 196, 219, 280, 332, 477).
Recycling Coenzyme A
The total CoA pool consists primarily of the nonesterified form (CoASH), and its thioesters acetyl-CoA, succinyl-CoA, and malonyl-CoA (214, 454). The size of the CoA pool is tightly regulated, remaining relatively constant, somewhere in the range of 100 to 500 μM (85, 454). This regulation occurs primarily through the utilization of pantothenate (the immediate CoASH precursor) and secondarily by degradation of CoASH (215, 216, 454). For reviews of CoASH biosynthesis, see references 41 and 213.
Because the CoA pool is limiting, its composition responds readily to the quality and quantity of the carbon source. This response is observed largely as a change in the ratio of acetyl-CoA to CoASH, whose concentrations vary inversely (84, 85). The nature of the carbon source affects this ratio. The addition to starved cells of assimilable carbon sources, e.g., d-glucose, causes this ratio to increase rapidly. In contrast, acetate, succinate, and nonassimilable sugars exert little or no effect on this ratio (85). This behavior explains, at least in part, why acetyl-CoA levels rise and then fall during growth on d-glucose and in tryptone broth (86, 360). The acetyl-CoA pool peaks during consumption of assimilable, acetogenic carbon sources and diminishes as the cells assimilate the previously excreted acetate (Fig. 1). Not surprisingly, this behavior correlates inversely with that of the TCA cycle, which becomes repressed during growth on d-glucose (6, 320, 342) and induced during growth on acetate (226, 329, 342).
Regenerating NAD+
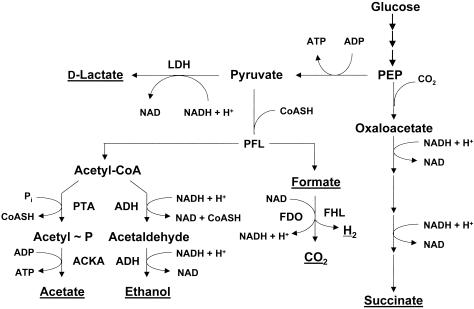
Glycolysis oxidizes glucose to two molecules of pyruvate while generating only two ATP molecules (Fig. 3). To fulfill their demand for ATP, therefore, E. coli cells must consume large amounts of glucose. Oxidation of glucose, however, also produces two molecules of NADH, which corresponds to four reducing equivalents. Because NAD+ serves as a substrate for the glycoytic enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH), cells must reoxidize NADH to maintain glycolytic flux. In the absence of a functional TCA cycle, they achieve this by placing the reducing equivalents onto partially oxidized metabolic intermediates, predominantly d-lactate, succinate, formate and ethanol, which cells excrete into their environment along with acetate (Fig. 4). Whereas acetate excretion generates energy in the form of ATP, excretion of these other metabolic intermediates sacrifices energy to consume reducing equivalents (89, 157, 326, 432; for a review, see reference 137).
FIG. 4.
Pathways for the excretion of partially oxidized metabolites. Excreted metabolites are underlined. LDH, lactate dehydrogenase; PFL, pyruvate-formate lyase; PTA, phosphotransacetylase; ACKA, acetate kinase; ADH, alcohol dehydrogenase; FDO, aerobic formate dehydrogenase; FHL, formate-hydrogen lyase.
Environmental Influences
The composition of excreted fermentation products depends on a number of environmental factors, including the oxidation state of the carbon substrate (3), the extracellular oxidoreduction potential (380), and the pH of the external environment (54, 241). For example, the oxidation state dictates the amount of NADH to be recycled and therefore the composition of the excreted fermentation products. The oxidation of glucose (oxidation state = 0) into two molecules of pyruvate produces two NADH molecules, each corresponding to two reducing equivalents. A more reduced sugar alcohol (e.g., sorbitol; oxidation state = −1) produces three NADH, while a highly oxidized sugar acid (e.g., glucuronic acid; oxidation state = +2) yields no NADH. Thus, to recycle the larger amount of NADH formed during growth on the more reduced sorbitol, cells must excrete more of the highly reduced ethanol (oxidation state = −2). In contrast, cells growing on glucuronic acid are redox balanced and thus do not need to produce ethanol. Instead, they can convert more of their pyruvate to acetate (oxidation state = 0) (3, 54). Similarly, external pH influences the composition of excreted products. Near or above pH 7, the predominant products are acetate, ethanol, and formate, with moderate amounts of succinate (43). As the pH drops, cells produce lactate instead of acetate and formate (68) and convert the formate to H2 and CO2 (386). Since oxidized sugar acids (e.g., gluconate, glucuronate, and galacturonate) provide much of the carbon available in the colon (5, 341) and the pH of the adult colon generally ranges between 6 and 8 (14, 55, 102), acetate is the major component of the excreted fermentation products.
Excretion Pathways
To excrete acetate (as well as formate and ethanol), E. coli cells must first decarboxylate pyruvate into acetyl-CoA. The conversion of pyruvate to acetyl-CoA can occur oxidatively under aerobic conditions and nonoxidatively under anaerobic conditions. Oxidative decarboxylation, a reaction catalyzed by the pyruvate dehydrogenase complex (PDHC), generates two additional NADH per glucose (Fig. 3 and 5). High concentrations of NADH inhibit PDHC activity (176). Thus, the PDHC does not operate under conditions, e.g., anaerobiosis, that do not favor the rapid reoxidation of NADH to NAD+. Note that anaerobiosis also represses transcription of the genes that encode the PDHC (362). Thus, oxidative decarboxylation functions primarily during respiratory metabolism, although some function may be retained during anaerobiosis (167, 223, 421).
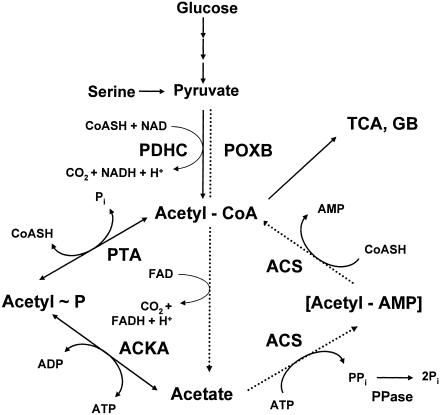
FIG. 5.
Acetate activation pathways. PDHC, pyruvate dehydrogenase complex; POXB, pyruvate oxidase; PTA, phosphotransacetylase; ACKA, acetate kinase; ACS, AMP-forming acetyl-CoA synthetase; PPase, pyrophosphatase; TCA, tricarboxylic acid cycle; GB, glyoxylate bypass. The dotted arrows denote the proposed PDHC bypass formed by POXB and AMP-ACS.
During anaerobiosis, cells of E. coli instead decarboxylate pyruvate to acetyl-CoA and formate by means of pyruvate formate-lyase (PFL), which catalyzes a nonoxidative reaction (242) (Fig. 4). Because its activity in vitro depends on an oxygen-sensitive glycyl residue, PFL has long been thought to function only in the absence of oxygen. However, more recent reports suggest that PFL can function in vivo in the presence of some oxygen (4, 113). This may occur through YfiD, which functions as a substitute glycyl radical domain to repair oxygen-induced damage to the PFL glycyl radical (461). The fate of the formate formed by PFL depends on the environmental pH. At neutral pH, formate is excreted. As the environmental pH decreases, depending on the availability of oxygen, formate decomposes either to carbon dioxide by aerobic formate dehydrogenase (FDO) or to both carbon dioxide and dihydrogen by formate-hydrogen lyase (FHL) (1, 4, 386).
The resultant acetyl-CoA follows two alternative fates: either conversion to acetate or reduction to ethanol. The conversion of acetyl-CoA to acetate, catalyzed by the PTA-ACKA pathway, generates two ATP molecules per glucose but consumes no reducing equivalents (Fig. 4). The reduction of acetyl-CoA to ethanol, catalyzed by alcohol dehydrogenase (ADH), sacrifices energy but consumes reducing equivalents. Thus, by modulating the amount of ethanol and acetate it excretes, a cell can balance its requirement to regenerate NAD+ with its need for energy (for a review, see reference 54).
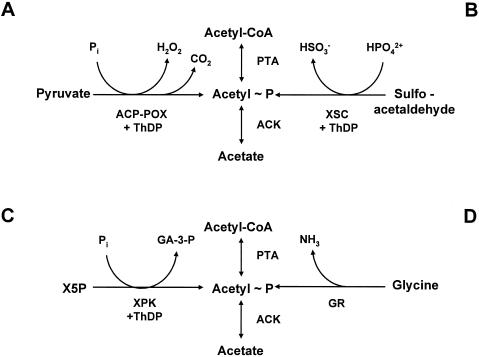
Acetate also can be excreted through the action of a third pyruvate-decarboxylating enzyme, pyruvate oxidase (POXB). Until recently, POXB had remained a mystery. This respiratory enzyme catalyzes the oxidative decarboxylation of pyruvate directly to acetate. The reaction produces carbon dioxide and reduces flavin adenine dinucleotide (FAD) (Fig. 3 and 5) (48, 49, 150). While the PDHC and PFL are considered essential enzymes, POXB generally has been regarded as nonessential and potentially wasteful (79, 80, 159). Mounting evidence, however, suggests that POXB provides energy and acetyl groups (as acetyl-CoA) under the microaerophilic conditions that prevail between exponential growth and stationary phase. Its transcription, dependent on σs, is induced in the early stationary phase. Although it is expressed maximally during aerobic growth, some expression does occur during anaerobic growth (80). POXB null mutants grow less efficiently than their wild-type parent. When overexpressed or expressed constitutively, POXB can substitute for the PDHC, albeit less efficiently. Thus, it has been proposed that POXB contributes substantially to aerobic growth efficiency (2). How does POXB perform this function? After prolonged incubation (in the absence of acetate), mutants that lack the PDHC form microcolonies, whose development depends on a functioning POXB (79). The simplest model has POXB and AMP-ACS forming a pyruvate-to-acetyl-CoA bypass of PDHC (2) (Fig. 3 and 5), a model easily tested by the construction of mutants that lack both the PDHC and AMP-ACS. Some existing evidence, however, is consistent with POXB and AMP-ACS functioning at the same time: cells appear to coordinate the induction of acs and poxB transcripts (347), while poxB mutants exhibit reduced acs transcription (249).
Acetate as an Acid
The acetate that cells excrete into the environment presents those cells with a problem. Acetate, like other weak acids, is toxic (280, 390). In its undissociated or acidic form, this lipophilic weak acid easily permeates membranes, uncoupling the transmembrane pH gradient (34, 35, 233, 375). Once across the membrane, it dissociates into a proton and an anion (56, 233). The proton acidifies the cytoplasm, while the anion increases the internal osmotic pressure and interferes with methionine biosynthesis (381, 382, 388). However, acetate is only partially oxidized and thus is a potential source of both carbon and energy. Therefore, the ability of E. coli to perform the “acetate switch” (61, 251) permits it to solve its acetate problem in a rather creative manner: it removes this potential toxin from its environment by consuming it.
ACETATE ACTIVATION PATHWAYS
During aerobic growth of E. coli on acetogenic carbon sources, the switch from dissimilation to assimilation requires two acetate activation pathways. Dissimilation depends primarily on the PTA-ACKA pathway, while assimilation functions primarily through AMP-ACS.
Acetate Dissimilation: the PTA-ACKA Pathway
In E. coli, acetate dissimilation is catalyzed by the enzymes PTA [acetyl-CoA(CoA):Pi acetyltransferase; EC 2.7.2.1] (294) and ACKA (ATP:acetate phosphotransferase; EC 2.3.1.8) (264). PTA reversibly converts acetyl-CoA and inorganic phosphate to acetyl∼P and CoASH, while ACKA reversibly converts acetyl∼P and ADP to acetate and ATP (Fig. 5) (385). Thus, the PTA-ACKA pathway couples energy metabolism with those of carbon and phosphorus (463, 465). This pathway also can interconvert propionyl-CoA and propionate (385). Thus, it also functions in α-ketobutyrate metabolism (457), degradation of fatty acids with odd numbers of carbons, the assimilation of propionate, and, in Salmonella, growth on 1,2-propandiol as a carbon and energy source (331).
Mutant phenotypes.
During shake flask growth, PTA-deficient mutants (pta or pta ackA) do not accumulate extracellular acetate (171, 172, 224, 360) (Table 1). In contrast, mutants that lack ACKA (ackA) accumulate small amounts of acetate (225, 360), which probably accumulates because the acetyl∼P produced by PTA is labile at physiological pH (61). During high-cell-density fermentations, pta or ackA mutants each accumulate a fraction of the acetate accumulated by their wild-type parent (78, 93, 172, 224, 483). However, their specific acetate production differs: ackA mutants produce acetate like their wild-type parents, while pta mutants exhibit a considerably smaller specific acetate production (93). Some evidence suggests that the small amount of acetate produced by pta mutants does not result from POXB (78), suggesting the involvement of another acetate-producing pathway.
TABLE 1.
Summary of pta and ackA mutant phenotypes relative to the wild-type
| pta mutant | ackA mutant |
|---|---|
| No or lowa excreted acetate | Lowb excreted acetate |
| Reduced specific acetate productiona | Wild-type specific acetate productionb |
| Increased pyruvate, lactate, and glutamate excretiona | NDc |
| Reduced formate and H2 excretiona | ND |
| Slow growth under acetogenic conditionsa | Slow growth under acetogenic conditionsa |
| No anaerobic growth on glucose | Anaerobic growth on glucose |
| Poor growth at high acetate concentration | Poor growth at high acetate concentration |
| Increased expression of the TCA cycle | Increased expression of the TCA cycle |
| Increased flux toward pyruvate | Increased flux toward pyruvate |
| Increased expression of YfiD | Increased expression of YfiD |
| Increased expression of acid resistance effectors | Increased expression of acid resistance effectors |
| Increased acid resistance | ND |
| Increased expression of chaperones and heat shock proteins | Increased expression of chaperones and heat shock proteins |
| Increased expression of some envelope proteins including OmpC | Increased expression of some envelope proteins but not OmpC |
| Decreased expression of some envelope proteins including OmpF | Decreased expression of some envelope proteins including OmpF |
| No acetyl∼P | Acetyl∼P accumulation |
| Excess flagella, especially at 37°C | Few flagella, especially at 37°C |
| Few pili | Many pili |
| Nonmucoid | Mucoid, even at 37°C |
| Poor survival during carbon starvation | Wild-type survival during carbon starvation |
During high-density fermentation.
During both high-cell-density fermentation and batch culture. All other phenotypes observed during batch culture only.
ND, not determined.
During aerobic growth on glucose, pta mutants excrete pyruvate, d-lactate, and l-glutamate instead of acetate, ethanol, formate, and dihydrogen (78, 118, 123, 224, 225, 482, 483). This resembles the normal behavior of wild-type cells exposed to low external pH, an environment that favors d-lactate excretion over that of acetate and formate. This behavior probably occurs because NADH-dependent lactate dehydrogenase (LDH) expression increases (68) while PTA expression and activity decreases (424, 437). Since wild-type cells exposed to high pH can tolerate a greater number of acid equivalents, they can up-regulate PTA expression (424), which favors the excretion of acetate (plus formate).
Excretion of d-lactate by pta mutants appears to replace excretion of ethanol as the primary mechanism for NAD+ regeneration. Excretion of l-glutamate is consistent with the observation that pta mutants up-regulate many TCA cycle genes (see below) and that aerobic growth on glucose represses KGDH (78, 439). The reduction in formate and dihydrogen levels suggests that the status of the PTA-ACKA pathway influences PFL activity (482), while the reduction in ethanol excretion supports the link between acetate (via the PTA-ACKA pathway) and ethanol (via ADH) as products of acetyl-CoA cleavage. Heterologous expression, in pta mutant cells, of enzymes or pathways that use acetyl-CoA as their substrate restores the wild-type pattern of fermentation products (with the exception of acetate) (78, 118, 482). These observations are consistent with the hypothesis that the PTA-ACKA pathway functions as an overflow pathway for excess carbon and that, in its absence, the cell uses alternative pathways to spill its excess carbon.
Cells that lack part or all of the PTA-ACKA pathway grow more slowly than their wild-type parents when grown aerobically in tryptone-based media, in defined minimal media supplemented with pyruvate, or in aerobic or anaerobic fermentations (61, 78, 93, 118, 172, 225, 240, 297, 466, 473, 482). In contrast, no defect occurs during growth on the nonacetogenic carbon source glycerol or the gluconeogenic carbon source succinate (78). The heterologous expression, in pta mutants cells, of enzymes or pathways that use acetyl-CoA as their substrate partially alleviates the growth defect (78, 118, 482). Thus, the growth defect might result from pyruvate accumulation, which would reduce the PEP/pyruvate ratio and, hence, lower the rate of substrate uptake by the PTS (78, 269, 340). Alternatively, it might be explained by a redox imbalance. Note that pta mutants cannot grow anaerobically on glucose unless they also lack ADH. These pta adh double mutants grow anaerobically on glucose by excreting the less reduced d-lactate via LDH instead of the highly reduced ethanol via ADH (Fig. 4). Furthermore, pta adh double mutants cannot grow anaerobically on sorbitol (a more reduced hexose than glucose) or glucuronate (a more highly oxidized hexose acid). Thus, the inability of pta mutants to grow anaerobically may result from redox imbalance (171). A similar imbalance might cause slowed aerobic growth of pta mutants, a problem that a heterologous acetyl-CoA-draining pathway could alleviate by depleting the acetyl-CoA pool and thus reducing the need to balance reducing equivalents.
Expression profile.
Slonczewski and coworkers (240) compared the proteome of wild-type cells to those lacking both PTA and ACKA. Similarly, Wolfe et al. (473) compared the transcriptomes of wild-type cells to those of mutants that lacked either ACKA alone or both PTA and ACKA. These analyses suggest that PTA-ACKA pathway mutants use a number of strategies in an attempt to cope with the loss of the energy-producing PTA-ACKA pathway. They increase the expression of TCA cycle enzymes, boost the expression of central glycolytic enzymes, and elevate the expression of a protein that facilitates PFL activity in the presence of oxygen (Table 1). In contrast, they do not increase the expression of the CoASH biosynthetic pathway or of enzymes involved in fatty acid biosynthesis, gluconeogenesis, or other fermentation pathways, including LDH and ADH. These mutants do, however, act as if they experience stress, especially that associated with the envelope and exposure to acid (240, 473). Indeed, their behavior resembles that of wild-type cells exposed to high acetate concentrations, an environment that results in down-regulated PTA expression (240).
Relative to their wild-type parents, both pta ackA and ackA mutants increase the steady-state transcript levels of genes that encode much of the TCA cycle (see Table S3 in the supplemental material of reference 473). They increase gltA (CS), icdA (IDH), sucD (a subunit of the SCSC), sucA (a subunit of KGDH), lpdA (a subunit of both the SCSC and PDHC), mdh, and b0725 (annotated as a hypothetical protein, but possibly only the 5′ untranslated portion of the sdh-suc transcript) expression. This response suggests that PTA-ACKA pathway mutants attempt to compensate by using the TCA cycle to recycle CoASH and to generate ATP and is consistent with l-glutamate excretion. When coupled with the knowledge that both glucose starvation and anerobiosis increase the expression of the PTA-ACKA pathway while decreasing that of the TCA cycle enzymes and subunits IDH, SUCC, SUCB, SDHA, and LPDA (324), this response implies that elevated carbon flux through the PTA-ACKA pathway somehow decreases the expression of the TCA cycle. Because pta mutants survive glucose starvation poorly while ackA mutants survive about as well as their wild-type parents, Nyström proposed that acetyl∼P might mediate this starvation response (324). However, heterologous expression in a pta mutant of an acetyl-CoA-draining pathway improves survival. Thus, Pan and coworkers have argued against Nyström's hypothesis (78). Interpretation of these studies remains problematic because of the intimate links between acetyl∼P synthesis and acetyl-CoA levels. Future studies might benefit from a molecular and/or genetic system that uncouples this relationship, i.e., a system that permits acetyl∼P synthesis without manipulating the acetyl-CoA pool or adding exogenous acetate, which can alter both the external and internal pH.
Both pta ackA and ackA mutants increase the steady-state level of the gapA transcript, which encodes GAPDH, the enzyme that catalyzes the NAD+-dependent oxidation of glyceraldehyde-3-P (Fig. 3). Similarly, pta ackA mutants increase the expression of triosephosphate isomerase, which catalyzes the interconversion of dihydroxyacetone phosphate and glyceraldehyde-3-P, just prior to the reaction catalyzed by GAPDH (Fig. 3) (240, 473). Note that both these mutants also up-regulate pfkB, which encodes the minor isoform of phosphofructokinase (473). Thus, pta ackA mutants may compensate by increasing carbon flux toward pyruvate. If so, then this compensation should increase the excretion of pyruvate and d-lactate, as observed (78, 482), and increase the amount of NADH.
PTA-ACKA- and ACKA-deficient mutants increase the steady-state levels of the YfiD protein and its transcript (240, 473). It has been proposed that YfiD acts as a substitute glycyl radical domain used by oxygen-stressed cells to repair oxygen-induced damage to the glycyl radical of PFL (461). Consistent with this hypothesis, growth in the presence of pyruvate or under microaerobic conditions elevates the level of YfiD (50, 290), while the pyruvate-sensitive regulator PdhR and the oxygen-sensitive global regulators FNR and ArcA control yfiD transcription (290, 476). Cells also up-regulate YfiD when exposed to low pH (50, 476) or to the organic acids propionate (50), benzoate (476), or acetate (240). Up-regulation also occurs in cells that express the heterologous FNR protein HlyX (161) or a heterologous acetyl-CoA-draining pathway (175). Intriguingly, peptidoglycan-free L-forms up-regulate YfiD, which appears as a phosphoprotein (141). In contrast, cells entrapped in gels down-regulate YfiD (344), as do those that overproduce threonine and excrete less acetate (262) or lack the global regulator FlhDC or the aerotaxis receptor Aer (356). Since YfiD is induced in acidic and/or microaerobic environments, it has been proposed to prevent the accumulation of acidic fermentation products by facilitating carbon flux through PFL (476). It remains unclear why PTA-ACKA pathway mutants attempt to compensate for their reduced ability to process acetyl-CoA by increasing the efficiency of an enzyme complex (PFL) whose products include acetyl-CoA. Perhaps these cells drive PFL backwards, thereby producing pyruvate and d-lactate instead of formate, as proposed by Slonczewski and coworkers to explain why formate results in reduced expression of acetate-induced proteins (424).
pta mutants resist extreme acid stress better than their wild-type parents do, a behavior that depends on crl, which encodes a transcription factor required for most σs-dependent phenomena (240). Consistent with their enhanced resistance to extreme acid stress, PTA-ACKA pathway mutants increase transcript and/or protein expression of Crl and a subset of σs regulon members (240, 473). This subset also includes a hyperosmotic stress-induced protein (OsmY), a stationary-phase-induced nucleoid protein (Dps) that contributes to acid and oxidative stress tolerance, and two periplasmic chaperones (HdeA and HdeB) induced by extreme acid stress. It also includes PFKB and the tryptophan repressor binding protein WrbA.
Transcripts of other genes implicated in stress responses also accumulate, most notably those that encode key chaperones and heat shock proteins (DnaK, GroEL, GroES, and ClpB) (473). The levels of most of these transcripts and/or proteins also become elevated in cells exposed to benzoate (256) or in those that express a heterologous acetyl-CoA-draining pathway (175). Other up-regulated stress genes include yhiE (which encodes a hypothetical protein implicated in the response to acid stress) and slp (encoding an outer membrane protein induced by carbon starvation). They also include ivy (formerly known as yfkE) (240, 473), whose gene product inhibits vertebrate C-type lysozyme.
In addition to HdeA and HdeB, increased expression of several other genes and/or proteins suggests that PTA-ACKA pathway mutants experience stress that affects the ability of the cell to fold, assemble, and/or insert envelope proteins. Such mutants up-regulate rseA, which encodes a negative regulator of σE, the sigma factor that responds specifically to periplasmic stress. They up-regulate RfbX, a putative O-antigen transporter. They also increase the expression of the outer membrane protease OmpT (240, 473). Finally, mutants that lack both PTA and ACKA, but not those that lack only ACKA, up-regulate the outer membrane porin OmpC (240, 473).
Several envelope- and/or stress-associated transcripts become down-regulated in PTA-ACKA pathway mutants (see Table S4 in the supplemental material of reference 473). These include genes that encode an outer membrane porin (OmpF), a component of the general secretory apparatus (SecG), a sugar-binding integral membrane component of a PEP-PTS system (GlvC, also known as PtiC), and a cytoplasmic, ribose binding component of the high-affinity ribose transport system (RbsD). They also include genes encoding a flagellar biosynthetic chaperone (FlgN), a hypothetical fimbrial chaperone (YqiH), and a capsule biosynthetic protein of unknown function (WcaM). Finally, they include genes that encode the protein repair enzyme isoaspartyl dipeptidase (Iad) and a cold shock protein (CspC) (240, 473). CspC stabilizes the rpoS transcript (346); thus, one might expect a decrease in the transcription of σs-dependent genes. Instead PTA-ACKA pathway mutants increase transcription of these genes, which argues that CspC does not play a key regulatory role under these conditions. Note that the two-component sensor RcsC negatively regulates yqiH, glvC and cspC. In contrast, it increases the expression of ivy and osmY (129), genes up-regulated in PTA-ACKA pathway mutants.
Acetate Assimilation: AMP-Forming ACS
In E. coli, AMP-ACS (acetate:CoA ligase [AMP forming]; EC 6.2.1.1) catalyzes acetate assimilation. A member of the firefly luciferase superfamily (445, 446), AMP-ACS first converts acetate and ATP to the enzyme-bound intermediate acetyladenylate (acetyl-AMP) while producing pyrophosphate (Fig. 5). It then reacts acetyl-AMP with CoASH to form acetyl-CoA, releasing AMP (46, 87). For a review concerning structure-function relationships, see reference 427.
Although reversible in vitro, this reaction is irreversible in vivo because of the presence of intracellular pyrophosphatases (PPase). This high-affinity pathway (Km of 200 μM for acetate), therefore, functions only anabolically, scavenging for small amounts of environmental acetate (61, 251). The reversible PTA-ACKA pathway also can assimilate acetate, but only in relatively large concentrations, because the enzymes of this low-affinity pathway possess Km values for their substrates in the 7 to 10 mM range (61, 251).
Because acetate freely permeates the membrane in its undissociated form (56, 233, 375, 390), assimilation does not require a dedicated transport system. However, under certain circumstances acetate uptake is saturable, suggesting that such a system exists (224). Recently, Gimenez et al. reported the existence of an acetate permease (ActP, formerly YjcG) and provided evidence for the existence of a second acetate transporter. The authors propose that these systems play critical roles when cells scavenge for micromolar concentrations of acetate (153).
Mutant phenotypes.
Cells that lack AMP-ACS (acs) grow poorly on low concentrations of acetate (≤2.5 mM) as their sole carbon and energy source (251). In contrast, cells defective for all or part of the reversible PTA-ACKA pathway grow poorly on higher concentrations of acetate (≥25 mM) (61, 224, 251). Since growth on low concentrations of acetate depends on AMP-ACS while growth on high concentrations requires the PTA-ACKA pathway, mutants that lack both cannot grow on acetate at any concentration (251). The inability to grow on acetate at any concentration argues against compensation by other acetate-activating enzymes, such as propionyl-CoA synthetase, encoded by prpE (201), or the propionate kinases, encoded by tdcD in E. coli or Salmonella enterica (186) or by pduW in S. enterica (52, 331). PrpE can use acetate as an alternative substrate (201); thus, PrpE can restore growth on acetate to acs pta ackA mutants, but only when expressed from a high-copy-number plasmid and then only poorly (S. Kumari and A. J. Wolfe, unpublished data). Similarly, AMP-ACS can activate propionate; thus, cells must lack both AMP-ACS and PrpE (acs prpE) before they can no longer grow on low concentrations of propionate (201).
In contrast to null mutants of acs, those that lack actP grow about as well as their wild-type parent on all concentrations of acetate tested. Gimenez et al. explain this puzzling result by noting that the lowest concentration tested (2.5 mM) is 50 times higher than the Km for ActP (5.4 μM). Since AMP-ACS possesses a Km of 200 μM for acetate, the authors propose that ActP functions with AMP-ACS to scavenge acetate at considerably lower concentrations than those tested (153). The observation that acs mutants compete poorly with their wild-type parent during prolonged starvation (A. J. Wolfe, unpublished data) is consistent with this proposal. Presumably, acs cells cannot scavenge for the small amounts of acetate released into the environment by dying cells. If so, then AMP-ACS (and perhaps ActP) should play critical competitive roles in any environment depleted of primary carbon sources.
When grown aerobically in shake flask culture, acs mutant cells grow as rapidly and accumulate as much acetate as their wild-type parents do (249, 251), exhibiting only a slight reduction in total biomass (A. J. Wolfe and C. M. Beatty, unpublished data). This reduction probably occurs because they cannot assimilate the previously excreted acetate even though they possess a fully functionally PTA-ACKA pathway (249, 251). Although reversible, the PTA-ACKA pathway generally does not participate in the assimilation of excreted acetate by cells grown in shake flask culture. This inability to participate in acetate assimilation arises because the extracellular acetate concentration generally does not exceed 1 mM and the enzymes of this low-affinity pathway possess Km values for their substrates in the 7 to 10 mM range (61, 251). In contrast, the PTA-ACKA pathway contributes to acetate assimilation during high-cell-density fermentation, probably because such growth results in considerably larger concentrations of extracellular acetate (93, 241, 455).
In glucose-controlled high-cell-density fermentation, the behavior of the acs mutant is puzzling. It grows initially with the same maximum specific growth rate as its wild-type parent. Growth slows, however, after a few hours and the culture achieves less than half the final biomass attained by the wild-type parent. Intriguingly, acs mutants accumulate only about a quarter as much extracellular acetate (1.8 g/liter, compared to 6.9 g/liter for the wild-type), while their specific production of acetate falls to about half. These results may implicate AMP-ACS in the control of carbon flux through the PTA-ACKA pathway (93). They also may indicate that AMP-ACS controls the expression and/or activity of the glyoxylate bypass (GB) of the TCA cycle (Fig. 3). During high-cell-density fermentations, the acs mutant achieves extracellular acetate concentrations high enough to permit assimilation by the low-affinity PTA-ACKA pathway (93). If the absence of AMP-ACS permits increased carbon flux through the GB, then the AMP-ACS-deficient mutant might accumulate extracellular acetate to a level lower than predicted by its specific rate of production. Indeed, two predictions of this model appear likely. First, a derivative of E. coli K-12 (JM109) accumulates considerably more extracellular acetate than does a derivative of E. coli B (BL21), although both produce acetate at the same specific rate (456). The GB appears to provide much of the difference. Flux and transcription analyses show that JM109 possesses a relatively inactive GB. In contrast, the GB of BL21 is considerably more active (323, 347). Second, acs mutants exhibit substantially elevated aceBAK transcription (Wolfe and Beatty, unpublished). Efforts to elucidate the relationship between the expression of AMP-ACS and that of the GB should help us understand why cells undergo the Crabtree effect.
Glyoxylate bypass and gluconeogenesis.
To assimilate acetate through either AMP-ACS or the reversible PTA-ACKA pathway, E. coli cells also require the GB (to replenish the TCA cycle) and gluconeogenesis (to provide sugar phosphates). Using two unique enzymes, isocitrate lyase (ICL) and malate synthase (MAS), induced during growth on acetate or fatty acids, the GB bypasses the two oxidative steps in the TCA cycle that evolve CO2 (Fig. 3). By avoiding the loss of two carbons at the expense of energy production, the GB permits the net accumulation of four-carbon biosynthetic precursors (e.g., succinate) during growth on two-carbon substrates (e.g., acetate) (for reviews, see references 90, 95, 100, and 246). Cells balance energy and biosynthetic needs, using exquisitely sensitive biochemical and genetic switches to control the flow of isocitrate through the TCA cycle and the GB. The biochemical switch modulates the activities of ICL and IDH (258). Whereas the phosphorylation of both ICL and IDH channels the flow of isocitrate into the GB, the dephosphorylation of both enzymes favors flow through the TCA cycle (203, 257). The genetic switch controls the aceBAK operon, which encodes ICL, MAS, and IDHK/P, the enzyme that reversibly phosphorylates IDH (44, 149). It involves repression by the GB regulator ICLR and activation by the nucleoid protein IHF and the global regulator Cra (also known as FruR) (reviewed in reference 95). ICLR also disassembles those transcription complexes that form, thereby augmenting its negative effect upon aceBAK transcription (478). For gluconeogenesis, PEP carboxykinase (PCKA) converts oxaloacetate (OAA) to PEP and the malic enzymes (encoded by sfcA and maeB) plus PEP synthase (PPSA) convert malate to PEP (329) (Fig. 3). Other gluconeogenic enzymes and reversible glycolytic enzymes drive the PEP toward glucose-6-phosphate. Note that Cra also activates genes that encode gluconeogenesis enzymes (319, 366, 367, 389).
Expression profile.
Relative to growth on glucose, wild-type cells grown on high concentrations of acetate (42 or 84 mM) up-regulate the steady-state levels of the transcripts and proteins of the following enzymes and pathways: AMP-ACS, the GB, the TCA cycle, and gluconeogenesis. In contrast, these cells down-regulate the transcripts and proteins of the PTA-ACKA pathway, nonacetate carbon transport systems, and glycolytic enzymes (226, 328, 329, 342, 451). Expression profiling of acs mutants has not been reported.
REGULATION OF THE ACETYL∼P POOL
The concentration of the intracellular acetyl∼P pool should depend on the following factors: expression and activity of the pathway components, and the availability of pathway substrates.
Expression and Activity of the PTA-ACKA Pathway
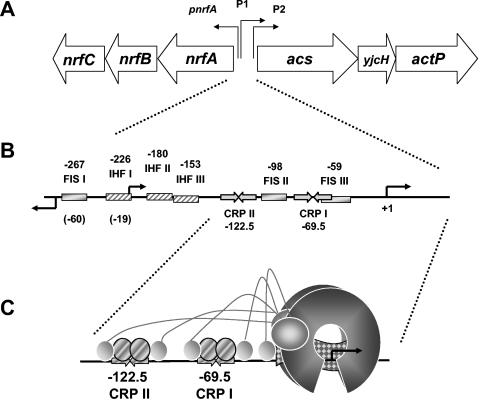
In E. coli and S. enterica, ackA and pta are contiguous (254, 466, 479). In E. coli, this operon produces two transcripts, one that encodes both ackA and pta and a second that encodes only pta (224). On the basis of genetic evidence, each transcript appears to result from a distinct promoter, one positioned upstream of ackA and another upstream of pta (466). In S. enterica, in contrast, the evidence suggests a single transcript driven by a single promoter (254).
In E. coli, the steady-state levels of ackA and pta transcripts and their gene products ACKA and PTA, as well as their enzymatic activity, vary in response to diverse environmental factors. Oxygen tension is a major influence. Under anaerobic conditions, the levels of ACKA and PTA increase 8- and 10-fold (324) but their activities increase only 2- to 3-fold (61). In S. enterica, ackA transcription increases in response to anaerobiosis, but only about 2-fold (254). As described previously, the environmental pH and the presence of acetate both exert sizeable effects. During aerobic growth, the steady-state level of PTA decreases as the environment acidifies and increases as the alkalinity rises (424). In contrast, during anaerobic growth, the steady-state level of ACKA increases as the environment becomes more acidic (220). During growth on or after exposure to high concentrations of acetate, the levels of PTA protein and of pta and ackA transcripts decrease about two- to threefold (240, 328, 329, 451). Temperature also exerts an effect: as it increases, so too does PTA activity. In contrast, ackA transcription decreases (360). The richness of the medium also plays a role: the steady-state levels of both PTA and ACKA, and of ackA transcripts, rise three- to fivefold in glucose-supplemented rich medium relative to glucose-supplemented minimal medium (324, 440). A similar effect occurs on starvation: ACKA and PTA levels increase 1.5- and 3-fold, respectively (324). The nature of the carbon source influences ACKA and PTA activities, but only marginally; they double during growth on pyruvate, d-lactate, or d-gluconate relative to growth on all other carbon sources tested, including d-glucose and acetate. The increased PTA activity probably results from the ability of pyruvate to activate the enzyme (437). In contrast, the carbon source exerts no apparent effect on the level of transcript or protein in either E. coli (324) or S. enterica (254). Finally, NADH, ADP, and ATP inhibit PTA activity (437). Thus, a warm, alkaline, anaerobic, and nutrient-rich environment generally favors increased pathway expression and/or activity while a cool, acidic, aerobic, and acetate-rich environment does not.
Availability of Pathway Substrates
The balance between intracellular acetyl-CoA and extracellular acetate probably represents the most important influence on the intracellular acetyl∼P pool. This should be especially true when the concentrations of extracellular phosphate, the total intracellular CoA pool, and the energy charge in ATP and ADP remain constant (196, 297). Under these conditions, the steady-state concentration of acetyl∼P should vary with the substrate (acetyl-CoA) and the product (acetate) and should be influenced by the same environmental factors that affect either. These factors include the acetogenic nature and amount of the carbon source, the availability of oxygen or nitrogen, the glycolytic flux, and the environmental pH and temperature.
Measurements of the Acetyl∼P Pool
Direct measurements of acetyl∼P in bacterial lysates have been reported on only a few occasions. Two biochemical approaches have been used: one that relies on two-dimensional thin-layer chromatography (53), and one that uses a coupled assay (208).
Prüβ and Wolfe, using an adaptation of Hunt's assay, observed a strong correlation between the pools of acetyl∼P and acetyl-CoA during growth in tryptone broth (Fig. 1B). Immediately following dilution of overnight cultures into fresh tryptone broth, acetyl∼P was undetectable (≤5 μM). The concentrations of both metabolic intermediates increased throughout the earliest stage of exponential growth, achieving a peak of 300 to 400 μM. The concentrations of both acetyl-CoA and acetyl∼P decreased progressively, reaching about 20 μM by entry into stationary phase; that of acetyl∼P remained low well into stationary phase (360). These results agree with measurements of acetyl-CoA levels in E. coli grown aerobically on glucose (86, 438). When one compares the consumption of amino acids with the accumulation of acetate and its activated derivatives, the following becomes apparent: acetyl-CoA and acetyl∼P levels peak during the transition from l-serine to l-aspartate consumption, while extracellular acetate peaks during the transition from l-aspartate to l-tryptophan consumption. The reaccumulation of extracellular acetate during stationary phase occurs as cells cometabolize acetogenic amino acids, e.g. l-threonine and l-alanine, with those that require the TCA cycle, e.g., l-glutamate.
Using the two-dimensional thin-layer chromatography approach, McCleary and Stock showed that acetyl∼P levels correlate with the acetogenic nature of the carbon source. Since the identity or quantity of the carbon source imposes only small effects on pathway activity and none on expression, these observations support the proposition that carbon flux exerts a major influence over the size of the acetyl∼P pool. If the sole carbon source was either pyruvate or d-glucuronate, cells grown in low-phosphate defined medium, and harvested at midexponential phase, possessed high concentrations of acetyl∼P (>1,000 μM). If the carbon source was d-glucose, they accumulated moderate amounts (300 μM). If the carbon source was d-fructose, glycerol, or fumarate, the levels were low or undetectable (≤50 μM). Relative to cells harvested during exponential growth, those harvested during early stationary phase increased their acetyl∼P pool three- to fourfold, but only if grown on d-glucose or d-fructose. If grown on pyruvate, in contrast, the pool decreased by two- to threefold (297). Note that cells grown on pyruvate behave quite similarly to cells grown in tryptone broth, i.e., elevated levels of acetyl∼P during exponential growth and reduced levels during stationary phase (297, 360). This should not be surprising, since cells convert l-serine, the preferred carbon source in tryptone broth (359), to pyruvate by means of an energy-independent transaminase reaction (334).
Prüβ and Wolfe also demonstrated a correlation between incubation temperature and the intracellular acetyl∼P pool. At or below 34°C, they could not detect acetyl∼P; above that temperature, the concentration increased. The influence of temperature became more obvious in mutant cells that lacked ACKA; acetyl∼P could be detected at 34°C, and its concentration rose dramatically up to 40°C (360). These results are consistent with the observations that extracellular acetate correlates with temperature (250) and can be readily explained by reduced ackA transcription coupled with increased PTA activity (360). Using a similar rationale, one might predict elevated acetyl∼P levels during growth on glycolytic intermediates at alkaline pH. Such an environment should favor synthesis of PTA over that of ACKA.
Indirect measures also have been performed, using acetyl∼P-dependent reporter systems. The best example interconnects nitrogen assimilation and acetyl∼P. In the absence of NRII, acetyl∼P acts as the predominate source of phosphoryl groups for the activation of the NRI and, hence, the transcription of glnA (Fig. 6). Since glnA encodes the enzyme glutamine synthetase (GS), one can monitor acetyl∼P concentrations indirectly by measuring GS activity. Using this assay, Atkinson and Ninfa demonstrated that the status of a cell's acetyl∼P pool depends on its ability to modify GS and regulate its activity. An adenylyltransferase (ATase) inactivates GS; thus, one would expect ATase-deficient cells to exhibit elevated GS activity. Instead, cells that lack both ATase and NRII display reduced GS activity. Since this activity depends on PTA, the ATase must regulate acetyl∼P levels. This regulation probably occurs indirectly by controlling flux through acetyl-CoA. Because l-glutamate, a GS substrate, is derived from 2-ketoglutarate, elevated GS activity may deplete TCA cycle intermediates. This would draw acetyl-CoA into the TCA cycle, diminish flux through the PTA-ACKA pathway, and hence reduce acetyl∼P accumulation. In contrast, reduced GS activity should favor elevated acetyl∼P levels (20, 127). Direct measurements should be performed to test these conclusions and those derived from other acetyl∼P-sensitive reporter systems.
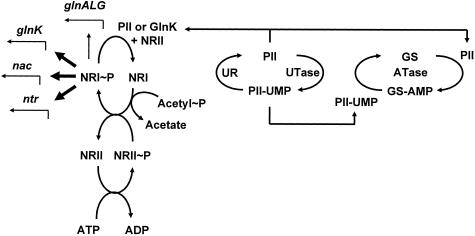
FIG. 6.
Regulation of nitrogen assimilation. Uridylyltransferase (UTase) and uridylyl-removing enzyme (UR) sense the relative amount of nitrogen. Under limiting conditions, UTase uridylylates PII to PII-UMP. In excess nitrogen, the UR deuridylylates PII-UMP to PII. PII favors adenylyltransferase (ATase) activity. ATase inactivates GS by adenylylating it to GS-AMP. PII (or its ortholog GlnK) also enhances the phosphatase activity of the histidine kinase/phosphatase NRII, which then dephosphorylates NRI∼P. This diminishes transcription from the nitrogen-regulated promoters such as ntr, nac, glnK, and glnALG. In contrast, PII-UMP enhances the deadenylylation of GS-AMP, hence activating the enzyme. PII-UMP exerts no direct effect on NRII; however, the lack of PII favors the kinase activity of NRII, which donates its phosphoryl group to NRI. Acetyl∼P also donates its phosphoryl group to NRI. Transcription from the glnALG promoter requires low amounts of NRI∼P (thin arrow), whereas transcription from the other promoters requires high amounts of NRI∼P (thick arrows).
ACETYL∼P AS A GLOBAL SIGNAL
Acetyl∼P is a high-energy form of phosphate (271). It possesses a larger ΔG° of hydrolysis (−43.3 kJ/mol) than ATP (−30.5 kJ/mol in complex with Mg+). Thus, acetyl∼P stores more energy than ATP and almost every other phosphorylated compound, with the notable exception of PEP (−62.2 kJ/mol) (265, 459). This is the basis for its pivotal role in ATP generation by substrate phosphorylation, as well as several proposed energy-related and signaling roles.
Energy Source for Transport
Acetyl∼P (or a compound derived from it) was proposed as the energy source for binding protein-dependent (BPD) ATP-binding cassette (ABC) transporters (197, 198). This proposal relied on metabolic inhibitors and mutants of the PTA-ACKA pathway to suggest a correlation between acetyl∼P and transport by BPD-ABC transporters. Strains that lack the pta-ackA operon are not defective for transport of nutrients accumulated by BPD-ABC transporters (90, 197), and studies with a reconstituted transport system in isolated membrane vesicles showed that acetyl∼P was not sufficient to energize transport (209). Given definitive evidence that ATP hydrolysis functions as the energy source, it appears that acetyl∼P does not play this proposed role. For a review, see reference 8.
Acetyl∼P also was proposed as an alternative to PEP as an energy source for the PTS (135). Working with purified proteins, Roseman and coworkers demonstrated a possible link between the PTA-ACKA pathway and the PTS. They showed that the purified phosphorylated form of EI of the PTS pathway can reversibly donate its phosphoryl group to purified ACKA and that this reaction permits the transfer of phosphate between EIIA of the PTS pathway and ACKA (135). Since acetyl∼P acts as a substrate for the phosphorylation of ACKA (136), the authors argued that acetyl∼P could function as an alternative phosphodonor for the uptake of PTS sugars. They proposed that such a transfer of phosphoryl groups could link the TCA cycle to the PTS, thereby permitting the energy charge (ATP/ADP ratio) and TCA cycle intermediates to regulate sugar transport (135). Since this reaction sequence has been demonstrated in vitro only, in vivo studies should be performed to determine whether this alternative pathway actually functions. If it does, then acetyl∼P could help to energize sugar transport during overflow metabolism or anaerobic growth on sugar, during which acetyl∼P accumulates.
Function in Signal Transduction
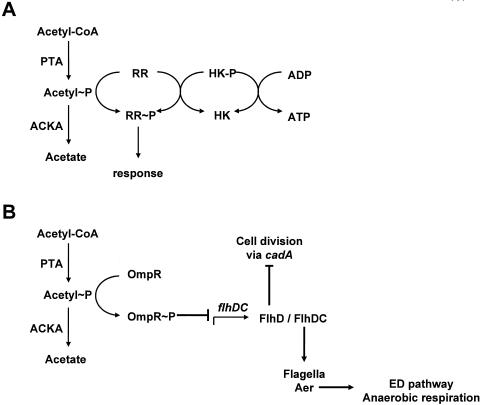
About a decade ago, two groups hypothesized that acetyl∼P functioned as a global signal (298, 463) and that it might act through its ability to donate its phosphoryl group to certain members of the family of two-component signal transduction (2CST) systems (127, 279). The most fundamental of these two-component signal transduction (2CST) pathways consists of a sensor and a response regulator (RR) (Fig. 7A). The sensor, a histidine protein kinase (HK), autophosphorylates a conserved histidine residue, using ATP (but never acetyl∼P) as its phosphoryl donor. The RR autophosphorylates on a conserved aspartate residue, using its phosphorylated cognate HK as its phosphoryl donor. Some HKs also possess phosphoaspartate phosphatase activity (hereafter called HKP), which may be their primary regulatory role (74, 279, 322). RRs also may be perceived as phosphatases, whose phosphorylated intermediate possesses a signaling function (393). Each of these phosphatases uses the phosphorylated form of its cognate HK as its substrate (298). Phosphohistidine phosphatases distinct from RRs also exist (292, 327, 378). For reviews, see references 148, 191, 429, 431, and 471.
FIG. 7.
(A) Acetyl∼P can donate its phosphoryl group to two-component RRs. HK-P, histidine kinase/phosphatase. (B) Model depicting how acetyl∼P can influence diverse cellular processes through the RR OmpR and its ability to repress transcription of flhDC. In contrast to acetyl∼P-dependent control of flagellar biosynthesis, its control of cell division does not require FlhC. ED, Entner-Doudoroff.
A wealth of data support the hypothesis that acetyl∼P can interact with 2CST pathways. In vitro, many RRs clearly autophosphorylate using acetyl∼P as their direct phosphoryl donor (Fig. 7A; Table 2). In vivo, however, the evidence for direct phosphorylation of RRs via acetyl∼P remains less certain. Most, but not all, reports that acetyl∼P can influence a RR-dependent behavior in vivo have relied on strains that lack the cognate HK (Table 2). Using this strategy, one manipulates acetyl∼P levels by varying the carbon source and/or by using mutants that lack PTA, ACKA, or both. The RR-dependent behavior is monitored indirectly by measuring the activity of a known target or reporter. These studies have demonstrated a strong correlation between the status of the acetyl∼P pool and the activation of some 2CST targets. Furthermore, an increasing minority of reports present evidence that acetyl∼P can influence the activity of certain RRs, even in the presence of the respective cognate HK (Table 2). Several mechanisms could explain this behavior. It could result from elevated ATP levels (264, 324, 343). It could indicate that a compound derived from acetyl∼P acts as the immediate phosphoryl donor (197, 198). It may reflect direct phosphorylation of a given RR, using acetyl∼P as the phosphoryl donor (127, 238, 297, 298, 360). Alternatively, an unidentified mediator might facilitate RR phosphorylation at the expense of acetyl∼P (127, 238, 264). A brief review of the evidence that acetyl∼P can serve as a phosphoryl donor in vitro is presented here, followed by a few examples that show the utility of the HK deletion strategy, provide evidence that acetyl∼P can influence the activity of certain RRs in vivo, and/or highlight salient outstanding issues. This is followed by evidence that acetyl∼P can indeed influence certain RR-dependent behaviors, even in the presence of the cognate HK. Finally, the merits of the proposed mechanisms are debated.
TABLE 2.
List of two-component RRs influenced by acetyl∼P
| RR | HK and/or HPT | Organism(s) | Presenta
|
Reference(s) | ||
|---|---|---|---|---|---|---|
| In vitro | In vivo (HK−) | In vivo (HK−) | ||||
| AlgR | AlgZ | Pseudomonas aeruginosa | + | 115 | ||
| BvgA | BvgS | Bordetella pertussis | + | 58 | ||
| CitB | CitA | Klebsiella pneumoniae | + | 304 | ||
| CheY | CheA | Escherichia coli | + | + | 106, 121, 279 | |
| CheY1, Y2 | CheA | Sinorhizobium meliloti | + | 417 | ||
| CheY1, Y3-6 | CheA | Rhodobacter sphaeroides | + | 128 | ||
| CheV2 | Helicobacter pylori | + | 348 | |||
| ComA | ComP | Bacillus subtilis | + | + | 237, 383 | |
| CpxR | CpxA | Shigella sonnei, E. coli | + | + | 107, 108, 110, 318b | |
| CsrR | CsrS | Streptococcus pyogenes | + | 47 | ||
| CyaC | CyaC | Spirulina platensis | + | 228 | ||
| DcuR | DcuS | E. coli | + | 217 | ||
| FixJ | FixL | Sinorhizobium meliloti | + | 376 | ||
| KdpE | KdpD | Clostridium acetobutylicum | + | 42 | ||
| KdpE | KdpD | E. coli | + | + | 182c | |
| MprA | MprB | Mycobacterium tuberculosis | + | 484 | ||
| NarL | NarX | E. coli | + | 268 | ||
| NodW | NodV | Bradyrhizobium japonicum | + | 275 | ||
| NRI (NtrC) | NRII (NtrB) | E. coli | + | + | + | 20, 127, 297, 360 |
| OmpR | EnvZ | E. coli | + | + | + | 24, 25, 181, 187, 273, 291, 297, 355, 408 |
| PhoB | PhoR | E. coli | + | + | 190, 264, 297, 466 | |
| PhoP* | PhoQ | Salmonella enterica | + | + | 76d | |
| PrrA | PrrB | R. sphaeroides | + | 92 | ||
| RcpA | CphA | Calothrix sp. strain PCC7601 | + | 205 | ||
| RcpB | CphB | Calothrix sp. strain PCC7601 | + | 205 | ||
| RegA | RdeA | Dictyostelium discoideum | + | 442, 443 | ||
| RssB | none | E. coli, S. enterica | + | + | 57, 103e | |
| SirA | BarA | S. enterica | + | 260f | ||
| SSK1 | SLN1-YPD1 | Saccharomyces cerevisiae | + | 218g | ||
| VanR | VanS | Enterococcus faecium | + | + | 18, 173, 194h | |
+, evidence exists.
In vivo (HK−) evidence supported by (i) differential effects during growth on pyruvate versus growth on glucose and (ii) behavior of pta ack mutant (ack mutant not tested).
In vivo (HK−) evidence supported by (i) behavior of pta ack mutant (ack mutant not tested) and (ii) mutagenesis found no additional factor.
PhoP* is an acetyl∼P-sensitive mutant. In vivo, wild-type PhoP does not respond to acetyl∼P.
Documented in E. coli, disputed in S. enterica.
In vivo (HK+) evidence supported by behavior of pta ack mutant (ack mutant not tested).
Poor in vitro phosphorylation enhanced by inclusion of phosphorylated E. coli CheY.
Acetyl-P hypothesized to activate VanR in vivo in E. faecium; in vivo shown to activate VanR expressed heterologously in E. coli that does not express the cognate HK VanS.
Evidence in vitro.
Many, but not all, RRs can use acetyl∼P as a phosphoryl donor to catalyze their own phosphorylation (Fig. 7A; Table 2). Other small phosphorylated compounds (carbamyl phosphate, phosphoramidate, monophosphoimidazole, and diphosphoimidazole) can perform a similar function, although no evidence exists that they perform this function in vivo. In contrast, neither ATP, PEP, nor creatine phosphate can donate their phosphoryl group to any RR (58, 115, 127, 190, 279, 296, 297, 376, 383, 411, 486). Each RR responds uniquely with respect to phosphoryl donor preference and its rate of autophosphorylation. Once phosphorylated, however, each RR behaves similarly by all criteria regardless of the phosphoryl donor, be it a small phosphorylated compound or a phosphorylated HK (127, 190, 279, 295, 297, 486).
Evidence in vivo. (i) Chemotaxis.
The first indication that an activated acetate might activate an RR in the absence of its cognate HK came from a study of the bacterial chemotaxis signal transduction pathway. Using E. coli cells that expressed the chemotaxis RR CheY, but not its cognate HK CheA, Wolfe et al. found that exogenous acetate increased the clockwise (CW) rotation of flagellar motors, a behavior that requires CheY. By varying the growth conditions, using metabolic inhibitors and mutants defective for CoASH biosynthesis and acetate metabolism, these authors concluded that CheA-independent CheY activation involved AMP-ACS and/or the activated acetate acetyl-AMP (474). Subsequently, Dailey and Berg discovered that the acs mutant used in the initial study also exhibited substantially reduced ACKA activity. They reexamined the acetate effect and found that it depended on the ability of the cell to synthesize acetyl∼P and the ability of CheY to accept a phosphoryl group, but did not depend on a functional PTS system (106). Later, it was shown that CheA-independent activation of CheY can occur by either of two mechanisms: (i) it can become phosphorylated using acetyl∼P as its phosphoryl donor, or (ii) it can become acetylated by AMP-ACS using as its substrate either acetyl-CoA or both acetate and ATP (26). Chemotaxis itself, however, does not require ACKA, PTA, or therefore acetyl∼P (106), most probably because the phosphotransfer from CheA to CheY is several orders of magnitude more efficient than the transfer from acetyl∼P (109, 295, 411). The role of AMP-ACS and acetyl-AMP in chemotaxis remains more controversial (see below).
(ii) Nitrogen assimilation.
A complex sensory system monitors nitrogen availability (Fig. 6). By favoring the phosphatase activity of the HKP NRII, this sensory system reduces transcription of the glnALG operon, which encodes GS, which interconverts l-glutamate and ammonia with l-glutamine. The operon also encodes a 2CST pathway, composed of NRII (also known as NtrB and encoded by glnL) and its cognate RR NRI (also known as NtrC and encoded by glnG). In contrast, limiting nitrogen favors the kinase activity of NRII, which autophosphorylates a conserved histidine residue and then donates that phosphoryl group to NRI. In addition, NRI can accept its phosphoryl group from acetyl∼P. Whereas small amounts of NRI∼P suffice for glnALG transcription (thin arrow in Fig. 6), larger amounts are necessary for the transcription of genes (ntr) that facilitate the utilization of secondary nitrogen sources (thick arrows). For full glnA transcription, phosphoryl group donation by either NRII-P or acetyl∼P suffices. In contrast, both donors are necessary for growth on certain secondary nitrogen sources, presumably because of the need for large amounts of NRI∼P. For reviews, see references 322 and 370.
Evidence supporting the participation of acetyl∼P in this regulatory scheme came from studies performed by Ninfa and coworkers. To examine the NRII-independent transcription of the glnALG operon, they constructed a nonpolar deletion of glnL, thus retaining an intact copy of glnG. They manipulated acetyl∼P levels by varying the carbon source or by using mutants lacking PTA or both PTA and ACKA. They monitored the activity of the RR NRI indirectly by measuring GS activity. They found that GS activity depended on NRI and, in the absence of NRII, correlated directly with the ability of cells to synthesize acetyl∼P. Since acetyl∼P can donate its phosphoryl group directly to purified NRI, they proposed that this might be the mechanism used by intact cells (127). Recently, this relationship was used by Laio and coworkers to engineer an artificial quorum sensory circuit (67).
Strong evidence supports the notion that the reverse is true, i.e., that nitrogen availability plays a key role in the regulation of intracellular acetyl∼P (20, 360). When harvested after several hours in stationary phase, cells that lack NRII (glnL) exhibit about eightfold-higher levels of acetyl∼P than do their wild-type parents, a behavior that occurs whether the cells have been grown in tryptone broth (TB), TB plus yeast extract (LB), or LB supplemented with a source of excess nitrogen (i.e., l-glutamine) (360). Since acetyl∼P can donate phosphate to NRI (127) and since NRII functions as an NRI∼P phosphatase under the conditions tested (322), this observation argues that NRII and NRI drain phosphoryl groups from acetyl∼P (20, 360). This hypothesis is consistent with the observation that glnL ackA double mutants grow poorly and accumulate suppressors while glnL pta ackA mutants do not (127). It remains to be seen whether glnL ackA mutants accumulate extremely elevated levels of acetyl∼P and, if so, how those elevated levels influence the growth rate.
(iii) Phosphate assimilation.
PhoR and CreC (formerly PhoM) are HKs that can donate their phosphoryl group to the RR PhoB (7, 288). When phosphorylated, PhoB activates transcription of the PHO regulon, including phoA, which encodes bacterial alkaline phosphatase (BAP) (287). Using the HK deletion strategy, Nakata and coworkers cloned the ackA gene serendipitously. Seeking the creC gene, they introduced a multicopy library of E. coli chromosomal fragments into a creC phoR strain (264) and screened for increased BAP activity. One such transformant carried ackA, which activated BAP expression only if overexpressed. Even then, the activity was considerably lower than that exhibited by wild-type cells or by phoR creC mutants that expressed creC from a plasmid. Since ackA overexpression also increased the intracellular ATP pool, the authors proposed that elevated ATP levels might facilitate autophosphorylation of a modulator protein that could cross talk to PhoB (264).
Subsequently, Wanner and Wilmes-Riesenberg showed that the induction of PhoR- and CreC-independent BAP activity depends completely on the ability to synthesize acetyl∼P. They noted that phoR creC mutants exhibited a BAP+ phenotype on energy-rich media and found that spontaneous and transposon-induced BAP+ suppressors of a phoR creC strain mapped to ackA. They manipulated the level of acetyl∼P by varying the carbon source and by introducing mutant alleles of pta or ackA or both. They monitored BAP activity and found that it correlated with the predicted acetyl∼P levels. They proposed two alternative mechanisms for acetyl∼P-dependent activation of PhoB: (i) indirect activation through a HK that senses acetyl∼P or (ii) direct activation, most probably by phosphorylation (464, 466). To distinguish between these two possibilities, they unsuccessfully sought mutants that lacked the acetyl∼P sensor (466). Instead, they identified additional mutants that influenced PhoR- and CreC-independent activation of the PHO regulon, including one with a mutation in the RR ompR (463). The subsequent attempt to understand this ompR effect highlighted several critical issues pertaining to the relationship between acetyl∼P and 2CST systems. First, it demonstrated clearly that acetyl∼P is responsible for PhoR-independent activation of the PHO regulon. It also showed that acetyl∼P-dependent activation functions independently of CreC-dependent activation and that the relative contributions of acetyl∼P and CreC depend on the carbon source and other characteristics of the culture medium. Second, it raised the possibility that E. coli possesses a PTA-independent mechanism for the synthesis of acetyl∼P, albeit one that produces only very small amounts. When grown in rich medium supplemented with glucose, phoR creC mutants that lack PTA exhibited 22.5 times more BAP activity than did those that lack both PTA and ACKA (238). Since these assays were performed during stationary phase, a PTA-bypass consisting of POXB and ACKA is a possible candidate (Fig. 5). Similar bypasses have been demonstrated or implicated in other organisms (343, 387). Third, it highlighted significant differences in the physiology and behavior of cells grown under different growth regimens. For example, phoR creC mutants exhibited substantial BAP activity on a minimal defined medium supplemented with pyruvate but not on the same medium containing glucose. These cells also exhibited significant BAP activity on a tryptone-based medium supplemented with glucose but not in the absence of that sugar (238). During exponential growth, metabolism of d-glucose, pyruvate, and the l-serine present in tryptone all lead to elevated acetyl∼P pools (297, 355, 360). As cells grown on d-glucose and pyruvate enter stationary phase, however, their behavior with respect to acetyl∼P pools diverges (297). Also, these BAP assays were performed with cells harvested from late stationary phase, and the little we know about the status of the acetyl∼P pool in stationary phase cells comes from early stationary phase. Thus, one must use caution when interpreting data obtained from stationary-phase cells grown with different carbon sources, at least until the relationship between the acetyl∼P pool and nonexponential growth conditions has been examined more thoroughly. Fourth, it exposed an intriguing connection between the PhoR/PhoB and EnvZ/OmpR 2CST pathways. phoR creC mutants that also lacked the RR OmpR, while retaining its cognate HK EnvZ, displayed 50 to 200 times more BAP activity on glucose and 2 to 3 times more activity on pyruvate. For glucose, this additional activity depended on the presence of EnvZ and the ability to synthesize acetyl∼P; for pyruvate, it depended on either. In all cases, EnvZ-dependent activation of PhoB required the absence of OmpR. Although the authors proposed several schemes to explain these observations (238), the mechanism by which acetyl∼P and EnvZ collaborate to facilitate PhoB phosphorylation remains unknown. Indeed, there is much that we do not understand about EnvZ and its cognate RR, OmpR.
(iv) Outer membrane porin expression and other OmpR-dependent behaviors.
The EnvZ-OmpR 2CST system mediates the regulation of the outer membrane porins, OmpC and OmpF (211). Although OmpR phosphorylation depends primarily on EnvZ, the following evidence supports the existence of an alternative pathway that involves acetyl∼P. Mutants that lack EnvZ retain some ability to phosphorylate OmpR (134) and hence can still regulate OmpC and OmpF expression (133, 394). In fact, the residual OmpF expression responds quite strongly to elevated osmolarity (133). The EnvZ-independent phosphorylation of OmpR depends, in large part, on the ability of cells to synthesize acetyl∼P (291). Even in cells that retain EnvZ, acetyl∼P can contribute to OmpR phosphorylation. In fact, acetyl∼P appears to play the predominant role when the environment becomes acidic (pH <6) (187) or depleted of nitrogen (273, 274), conditions under which EnvZ kinase activity remains low (187, 273). Similarly, the OmpR-dependent induction of acid tolerance in S. enterica does not depend on EnvZ but, rather, on the ability to synthesize acetyl∼P (24). In contrast, both EnvZ and acetyl∼P appear necessary for the autoinduction of ompR (25), a requirement similar to that for transcription of ntr genes (322).
A series of studies firmly established that OmpR can control flagellar biosynthesis in an acetyl∼P-dependent manner. First, Prüß and Wolfe demonstrated an inverse correlation between acetyl∼P levels and flagellar expression (360), confirming the observation that mutations in pta or ackA affect flagellar expression (409). Cells lacking PTA are hyperflagellated; in contrast, those lacking ACKA are poorly flagellated (360, 408). This led to the hypothesis that acetyl∼P donates its phosphate to a RR that represses transcription of the flhDC operon, which encodes the master flagellum-specific activator (360). Subsequent studies demonstrated that OmpR could directly repress flhDC transcription in an acetyl∼P-dependent manner, despite the presence of its cognate EnvZ (355, 408). Like PTA-deficient mutants, cells lacking OmpR are hyperflagellated (408). These observations led to the following model (Fig. 7B). Cells growing exponentially on an acetogenic carbon source possess elevated levels of acetyl∼P. Consequently, acetyl∼P acts a phosphoryl donor for OmpR, which then binds to and represses transcription from the flhDC promoter. Depletion of the acetogenic carbon source leads to reduction of the acetyl∼P pool, the accumulation of nonphosphorylated OmpR, and, thus, the derepression of flhDC transcription and flagellar expression (355, 358). This model makes some strong predictions, which can be tested directly. Since flagellation depends, in part, on the status of the acetyl∼P pool, manipulation of that pool should affect flagellation. Additional factors that could be tested include the nature and amounts of the carbon and nitrogen sources, the status of ATase, and osmolarity.
This model also can explain why the size of the acetyl∼P pool correlates with the rate of cell division. Cells in the exponential growth possess a large acetyl∼P pool and divide rapidly. As the carbon source becomes depleted, the acetyl∼P decreases, and the cells divide more slowly. Like repression of flagellar biosynthesis, this cell division behavior requires PTA, OmpR, and FlhD (but not FlhC). Mutants lacking FlhD continue to divide rapidly as they enter stationary phase, while those lacking PTA or OmpR cannot divide rapidly at all and instead form filaments (355, 358).
FlhD, alone or in combination with FlhC, acts a global regulator. In addition to regulating flagellar biosynthesis and cell division, it coordinates anaerobic respiration and the Entner-Doudoroff pathway, two responses critical to life in the colon (356, 357). FlhDC also regulates virulence factors in diverse organisms (51, 60, 132, 140, 235, 272, 399). Since acetyl∼P also contributes to OmpR autoinduction, it may help regulate the larger subset of OmpR-dependent yet FlhD-independent genes (330), including many associated with virulence (e.g., ssrAB, which encodes a 2CST pathway in S. enterica [261]). Indeed, acetyl∼P and ArcB provide OmpR with the phosphate necessary to induce the protective response to the neutrophil bactericidal/permeability-increasing (BPI) protein (354) (P. Prohinar and J. Weiss, personal communication).
(v) Implications for biofilm development and pathogenesis.
Acetyl∼P may play an even more global role, however, by influencing genes regulated by RRs other than OmpR. Recently, Wolfe et al. demonstrated that acetyl∼P can influence the in vivo expression of almost 100 genes (473). These authors compared the transcriptome of wild-type cells to those of cells that either accumulate acetyl∼P (ackA) or do not synthesize it (pta ackA). They designed this study specifically to differentiate genes that respond to perturbations in the acetyl∼P pool from those that respond to a general defect in the PTA-ACKA pathway, i.e., the inability to properly recycle acetyl-CoA, to generate ATP, or to excrete acetate. This study verified that acetyl∼P correlates with decreased expression of genes involved in flagella biogenesis (360) and showed that acetyl∼P correlates with increased expression of genes involved in type 1 pilus assembly, the biosynthesis of colanic acid (an extracellular polysaccharide or capsule), and the response to multiple stresses (Fig. 8).
FIG. 8.
Acetyl∼P-responsive processes and the RRs known to regulate them.
A number of RRs, other than OmpR, regulate the expression of these acetyl∼P-responsive genes (Fig. 8). Like OmpR∼P, RcsB∼P represses flagellar synthesis directly (138, 330). In addition, RcsB∼P activates both colanic acid biosynthesis (433) and at least one acetyl∼P-responsive stress effector (osmC) (112). Several other acetyl∼P-responsive genes are regulated by RcsC, the cognate HK of RcsB, suggesting that they might also be regulated by RcsB. These include ivy, osmB, osmY, yjbE, yiaD, and ycfJ. RcsC also regulates both rcsB and fimZ (138). FimZ∼P represses flagellar synthesis and activates type 1 pilus expression (91). UvrY (known as SirA in S. enterica and GacA in Pseudomonas aeruginosa) also represses flagellar expression (156). However, the mechanism remains obscure: its only known direct target is the regulatory RNA csrB (436), whose binding protein CsrA controls flagellar expression in a csrB-independent manner (467). Other RRs also control flagellar expression: ArcA and AtoC both activate it, while CitB and YpdB both repress it (330). It is not known which of these RRs function directly and which ones act indirectly, nor is it known which of these RRs mediate acetyl∼P-dependent regulation of flhDC or other flagellar regulators, e.g., the flagellum-specific sigma factor FliA.
Previously, other researchers had implicated flagella, type 1 pili, capsule, and diverse stress effectors in the formation of biofilms. Thus, Wolfe et al. tested whether cells altered in their ability to metabolize acetyl∼P could construct normal biofilms and found that they could not. Cells defective for the degradation of acetyl∼P or those defective for the production of acetyl∼P each could form biofilms; however, these biofilms displayed characteristics substantially different from each other and from the biofilms formed by their wild-type parent. The authors hypothesized that acetyl∼P helps coordinate expression of surface structures and cellular processes involved in wild-type biofilm development (473). Intriguingly, the ordered formation and dissolution of intracellular biofilm-like pods by uropathogenic E. coli depends on the very same structures that respond to the status of the acetyl∼P pool, i.e., flagella, type 1 pili, and capsule (10).
Most human pathogens, including E. coli, S. enterica, Vibrio cholerae, Yersinia pestis, B. anthracis, group A Streptococcus, and P. aeruginosa, possess pta and ack homologs. These pathogens also express RRs known to regulate virulence factors. In E. coli, acetyl∼P regulates the biosynthesis of several known virulence factors (473), and through its ability to control transcription of the flhDC operon and ompR (25, 356, 357), it probably regulates many more. In S. enterica, acetyl∼P can influence expression of virulence factors through SirA (260) but not induction of the pathogenicity island SPI-2 (234). Some evidence suggests a relationship between acetyl∼P and PhoP, although it probably does not influence gene expression in S. enterica (77). In V. cholerae, pta mutants do not produce the toxin-coregulated pilus, colonize mice poorly, and exhibit reduced virulence. Since overexpression of ToxT bypasses the pta defect, PTA probably sits above ToxT in the regulatory pathway. Because ToxT lacks a phosphate acceptor site, it probably does not receive phosphate from acetyl∼P. It is more likely that an RR exists that receives a phosphoryl group from acetyl∼P and, when phosphorylated, regulates either toxT transcription or the stability of ToxT. Alternatively, the RR∼P might help ToxT regulate its target genes (83). It also is conceivable that the lack of virulence results from the inability of the pta mutant to efficiently generate ATP, as proposed for hydrogen peroxide resistance in Streptococcus pneumoniae (343) (see below).
(vi) Implications for metabolic engineering.
Acetyl∼P also may impact efforts to engineer metabolic pathways. For example, the ability of cyanobacteria to accumulate polyhydroxybutyrate (PHB) depends, in part upon the synthesis by PTA of acetyl∼P, which activates PHB synthase. In cyanobacteria, PHB accumulates under conditions of excess acetate, excess reducing power, or deprivation of nutrients, such as nitrogen (19). Cells grown in nitrogen-sufficient conditions accumulate small amounts of PHB and exhibit low activities of both PTA and PHB synthase. In contrast, cells grown in nitrogen-limiting conditions accumulate large amounts of PHB and exhibit high PTA and PHB synthase activities. When added to crude lysates and membrane fractions isolated from cells grown under nitrogen-sufficient conditions, acetyl∼P activates PHB synthase by some, as yet unidentified, post-translational mechanism (308). This mechanism may be a general feature of polyhydroxalkanoate synthases, because acetyl∼P activates purified Thermus thermophilus PHA synthase (333). Surprisingly, pta mutants display enhanced PHB accumulation (309, 310). The authors explain this dichotomy, arguing that increased flux through acetyl-CoA in the pta mutant can compensate for the lack of acetyl∼P-dependent activation of PHB synthase (309). Taken together, these observations suggest that the concentration of acetyl∼P depends on the balance between nitrogen and carbon, that PHB synthase senses that balance by monitoring the acetyl∼P pool, and that efforts to increase the acetyl-CoA flux can override the need for acetyl∼P (308, 309).
How Does Acetyl∼P Work?
An increasing number of reports rely either explicitly or implicitly on the “fact” that acetyl∼P acts as a phosphodonor for RRs in vivo. That acetyl∼P influences gene expression appears solid enough; yet, the mechanism remains elusive and therefore controversial. Several mechanisms have been proposed to explain the link between acetyl∼P and gene expression. First, it might function indirectly by means of its connection to the PTS pathway (135). Second, it might increase energy metabolism, generating ATP by substrate phosphorylation (264, 324, 343). Third, it might bind ATPase and increase its activity. Fourth, it might directly donate phosphate to a RR in an HK-independent manner (127, 238, 297, 298, 360). Finally, it might facilitate the ability of an HK to donate a phosphoryl group to a noncognate kinase (127, 238, 264).
The most obvious connection of acetyl∼P to energy production depends on the presence of ACKA, the enzyme that interconverts acetyl∼P and ADP with acetate and ATP. Thus, acetyl∼P clearly can exert a global effect on energy in wild-type cells by enhancing ATP levels. Indeed, this appears to be the case in S. pneumoniae, where resistance to hydrogen peroxide appears dependent on the generation of ATP via acetyl∼P (343) (see below). However, the vast majority of in vivo experiments whose results support a signaling role for acetyl∼P use mutants that lack either ACKA or both ACKA and PTA. Thus, neither of these mutants can produce ATP via ACKA-dependent substrate phosphorylation. For the same reason, they cannot donate phosphate to EI of the PTS system (135). Thus, these mechanisms cannot explain the effects of acetyl∼P on gene expression, at least not in cells that lack ACKA.
Acetyl∼P could instead affect ATPase activity directly. The ackA mutant possesses elevated acetyl∼P levels (297, 360). It is formally possible that this excess acetyl∼P could bind ATPase, resulting in increased activity, which would not occur in the acetyl∼P-less pta ackA mutant. Indeed, such an interaction has been shown in vitro with certain mammalian ATPases (229, 415). To our knowledge, however, no evidence exists for such an interaction with bacterial ATPase. However, if acetyl∼P interacted efficiently with a bacterial ATPase, the concomitant increase in ATP synthesis might exert a stronger effect on certain 2CST pathways. Although all HKs use ATP as their phosphodonor, some may possess a higher Km and thus be more responsive to rising ATP levels. This hypothesis might explain some acetyl∼P-dependent effects; however, it cannot account for those observed in the absence of the cognate HK, unless cross talk with an alternative HK is invoked. Nevertheless, this hypothesis should be tested directly. Most importantly, it must be determined whether treatments that alter the ATP pool similarly influence acetyl∼P-sensitive 2CST pathways.
Another controversy revolves around the relationship between estimates of the acetyl∼P concentration in vivo relative to the concentration required for efficient phosphorylation in vitro. Clearly, the in vivo concentration of acetyl∼P (20 μM to 1.2 mM) is lower by several orders of magnitude than the concentration required to autophosphorylate most RRs in vitro (109, 295, 296, 411), an observation that some researchers have used to argue against a direct role for acetyl∼P as a phosphoryl donor (109, 238). Unfortunately, this argument is based on studies of PhoB and CheY, two RRs that appear unresponsive to acetyl∼P in the presence of their cognate HKs. Of equal importance, this argument fails to account for the consequences of molecular crowding. The inside of a cell is a very crowded environment, as crowded as a solution of 13% dextran (278). This environment is quite different from the dilute aqueous solutions generally used to study enzymatic reactions in vitro. The biochemistry of crowded solutions differs from that which operates in dilute solution (277, 278). For example, molecular crowding can slow protein diffusion, promote weak or unfavorable intermolecular associations, or drive unfavorable conformation changes. Thus, molecular crowding may cause the RR-acetyl∼P interaction to occur more readily than it does in dilute solution. For example, molecular crowding could induce a conformational change in an RR, poising it so that the acetyl∼P concentration required in vivo may be considerably lower than that required in dilute solutions. Thus, future in vitro kinetic studies should be performed in the presence of a crowding agent on RRs, e.g., OmpR or NRI, that readily respond to changes in the acetyl∼P pool.
It appears, therefore, that the “direct phosphodonor” model best explains most of the existing data. Most, but not all, of the connections predicted by this hypothesis have been documented, and no new players or mechanisms need to be found or envisioned. In vitro, acetyl∼P can clearly donate phosphate to a large subset of RRs. In vivo, about 100 genes respond to the status of the acetyl∼P pool, and many of these acetyl∼P-responsive genes are known to be regulated by a subset of RRs, including OmpR, NRI, and RcsB. Unfortunately, the final, critical in vivo connection has not yet been demonstrated: that a specific response occurs because acetyl∼P donates its phosphate to a specific RR. This demonstration is nontrivial. Only once has a phosphorylated RR been demonstrated in vivo (134). However, even this technically challenging procedure cannot distinguish an acetyl∼P-dependent phospho-RR from a HK-dependent phospho-RR. Remember that ATP represents the source of phosphate for both. Consequently, genetics remains the only recourse. If acetyl∼P donates phosphate to a specific RR, then an acetyl∼P-insensitive derivative of that RR, one that can accept phosphate from its cognate phosphoryl donor but not from acetyl∼P, should confer some subset of phenotypes that differs from those conferred by its wild-type parent. Fortunately, such an acetyl∼P-insensitive OmpR mutant exists (448), and it fails to mediate the OmpR-dependent response to the bactericidal/permeability-increasing protein (354). Preliminary studies, using this acetyl∼P-insensitive mutant (which exerts only limited effects on the canonical OmpR targets ompF and ompC), suggest that the ability of OmpR to influence biofilm development does not require EnvZ but does require that it receive a phosphoryl group from acetyl∼P (Wolfe, unpublished).
Clearly, some inconsistencies remain, the most pressing being the observation that the acetyl∼P-dependent activation of PhoB depends on EnvZ, at least under certain environmental conditions (238). What is the nature of the mechanism by which this noncognate HK influences the ability of PhoB to receive phosphate from acetyl∼P? It is highly unlikely that EnvZ autophosphorylates by using acetyl∼P as its phosphoryl donor. No reaction like this has ever been documented. As proposed by Wanner and colleagues, it is conceivable that EnvZ or some other HK senses acetyl∼P, autophosphorylates in response using ATP as its phosphoryl donor, and then donates that phosphoryl group to PhoB (238). Alternatively, EnvZ-P might donate phosphate to an unidentified RR, whose action combines with that of PhoB∼P (derived from acetyl∼P) to activate PhoA transcription. This scenario is somewhat analogous to that of growth on certain secondary nitrogen sources. There, NRI∼P must receive phosphoryl groups from two donors, NRII-P and acetyl∼P (322). Finally; EnvZ, in the absence of OmpR, might indirectly increase the acetyl∼P pool to a level that increases the phosphorylation of PhoB via acetyl∼P, a scenario that resembles the connection between GS and acetyl∼P (322).
What Does Acetyl∼P Signal?
An ideal signal of transient events possesses a short half-life and varies over a wide range of concentrations in response to those events. Acetyl∼P is the most unstable phosphorylated compound in the cell, and the size of its pool can vary at minimum 100-fold (360). The status of that pool appears to depend on the nature and amount of the carbon source balanced against the availability of oxygen, nitrogen, and phosphate and against the status of the TCA cycle (196, 297, 360, 463). Other environmental factors appear to impact the acetyl∼P pool: temperature, pH, and the concentration of extracellular acetate. Thus, the acetyl∼P pool could integrate information concerning the surrounding environment, e.g., the mammalian intestine (298, 321). Although the vast majority of that acetyl∼P donates its phosphate to generate ATP, a fraction is bled off to signal the network of two-component signaling pathways.
Two-component pathways tend to be present at low levels in unstimulated cells. In response to a specific stimulus, many of these pathways amplify themselves via positive autoregulatory loops. By donating its phosphate to the small amounts of selected RRs in unstimulated cells, acetyl∼P could “prime” whole subsets of pathways. Such “priming” could favor a more rapid transition to the activated state (321). For example, acetyl∼P might facilitate the transition from nitrogen-rich to nitrogen-poor environments (127). Relative to cells that lack it, those that possess NRII rapidly change their rate of glnA transcription after a nitrogen shift-up or shift-down (371). The slower response that occurs in the absence of NRII is controlled by acetyl∼P (127). Cells under nitrogen-rich conditions possess only a few molecules of NRII. As cells become limited for nitrogen, decreased glutamate synthase and glutamate dehydrogenase activity would limit the drain of 2-ketoglutarate from the TCA cycle. The diminished capacity of the TCA cycle should result in elevated acetyl∼P levels. Because glnL is part of the glnALG operon, the nitrogen status regulates NRII expression. Thus, phosphorylation of NRI by acetyl∼P could stimulate glnALG transcription, which would lead to increased amounts of both NRII and NRI. Now, the cell is “primed” to use both acetyl∼P and NRII-P to activate NRI for growth on secondary nitrogen sources. For reviews, see references 298, 321, and 322.
Characteristics of RRs That Respond to Acetyl∼P In Vivo
The ability of the acetyl∼P pool to inform RRs may represent a “primordial” mechanism that was later supplanted or appended by the evolution of HPKs (298). It is the latter class of RRs that are most likely sensitive to acetyl∼P. Although currently their number is small (Table 2), the RRs known (or suspected) to respond to acetyl∼P in vivo under physiologically relevant conditions seem to possess the following traits in common: (i) their cognate HKP functions primarily as a phosphatase, (ii) they exist in great excess over their cognate HK or HKP, or (iii) they seem to lack a cognate HK. For example, cells synthesize the RR OmpR in large excess over its cognate HK EnvZ (73), NRII functions primarily as an NRI∼P phosphatase (322), and the RR RssB (also known as SprE and MviA) possesses no specific cognate HK (183). These conditions represent opportunities for regulation via an HK-independent pathway, such as that envisioned for acetyl∼P. Clearly, further study is warranted.
Other Acetyl∼P-Forming Enzymes
In addition to PTA and ACKA, other enzymes evolve acetyl∼P. These include acetyl∼P-forming pyruvate oxidase,sulfoacetaldehyde acetyltransferase, fructose-6-phosphatephosphoketolase, and protein C of glycine reductase. These enzymes are found in diverse bacteria but not in E. coli and its relatives.
Pyruvate oxidase.
Pyruvate oxidases can be divided into two classes: the acetate formers such as POXB of E. coli and the acetyl∼P-formers (APF-POX [EC 1.2.3.3]) found in diverse lactobacilli, Pediococcus pseudomonas, and S. pneumoniae (49, 188, 315, 402, 403, 419). In the presence of inorganic phosphate, the thiamine diphosphate (ThDP)-dependent APF-POX catalyzes the oxidative decarboxylation of pyruvate to acetyl∼P, carbon dioxide, and hydrogen peroxide (Fig. 9A) (188, 315, 402, 403, 444). In S. pneumoniae, ACP-POX appears responsible for formation of the majority of the acetyl∼P, despite the presence of both ACK and PTA. ACP-POX-deficient mutants (spxB) generate dramatically reduced amounts of acetyl∼P (343) and produce undetectable levels of hydrogen peroxide (343, 419). spxB mutants are more sensitive to hydrogen peroxide exposure than are their wild-type parents, apparently because they cannot mount an acetyl∼P-dependent response. Because this response does not depend on de novo protein synthesis, it has been proposed that it does not involve the acetyl∼P-dependent induction of a stress effector (343). Instead, it may involve the ability of acetyl∼P to maintain the ATP pool. Exposure to oxygen or hydrogen peroxide causes the spxB mutant to lose substantially more of its ATP pool than wild-type cells do. Since the acetyl∼P pool exceeds the ATP pool by about 10-fold, the authors propose that ACP-POX and ACK form a hydrogen peroxide-resistant ATP-generating pathway, which provides energy when oxidative stress damages sugar transport and glycolysis, the primary sources of ATP for this strictly fermenting bacterium (343). Alternatively, acetyl∼P could regulate the stability of a previously expressed stress effector by modulating an RR, e.g., RssB, that regulates a protease activity (57).
FIG. 9.
Other acetyl∼P-forming enzymes. (A) ACP-POX, acetyl∼P-forming pyruvate oxidase; ThDP, thiamine diphosphate. (B) XSC, sulfoacetaldehyde acetyltransferase. (C) XPK, xylulose 5-phosphate phosphoketolase; GA-3-P, glyceraldehyde-3-phosphate. (D) GR, glycine reductase.
Unless supplemented with acetate, chemically defined medium does not support the aerobic growth of the spxB mutant (419). Thus, growth requires acetyl∼P (produced through either ACP-POX or ACK), which is then converted to acetyl-CoA by PTA (419). spxB mutants are poorly virulent, perhaps because they fail to adhere to the host cell glycoconjugates proposed to be components of pneumococcal receptors. The addition of acetate restores adherence, consistent with the idea that adherence and hence virulence, requires acetyl∼P (419). It is unclear, however, whether the decreased virulence of spxB mutants actually results from reduced acetyl∼P-dependent adherence. It also is unclear whether reduced virulence results from an inability to phosphorylate an RR, generate ATP under conditions of oxidative stress, or even generate hydrogen peroxide.
Sulfoacetaldehyde acetyltransferase.
Sulfoacetaldehyde is a key intermediate in the dissimilation of sulfonated compounds. For 30 years, it was believed incorrectly that the desulfonating enzyme cleaved sulfoacetaldehyde to acetate and sulfite (94). Recently, Cook and coworkers purified this desulfonating enzyme from several bacteria, demonstrated definitively that it actually produces acetyl∼P and sulfite, and renamed it sulfoacetaldehyde acetyltransferase (XSC) (EC 2.3.1.-) (Fig. 9B). This ThDP-dependent enzyme appears to be widespread. Bacteria from several subdivisions of Proteobacteria and at least a couple of gram-positive bacteria either exhibit XSC activity, express the protein, or possess the gene; they include Alcaligenes degragrans, Achromobacter xylosoxidans, Desulfonispora thiosulfatigenes, Sinorhizobium meliloti, Rhodobacter capsulatus, Alcaligenes defragrans, Paracoccus denitrificans, Rhodococcus opacus, and Ralstonia spp. (65, 114, 387). However, it is not present in E. coli (387). It is hypothesized that during fermentation or effective sulfite reduction, the acetyl∼P formed by XSC possesses two divergent metabolic fates: either conversion by PTA to acetyl-CoA for anabolism or as a phosphodonor for the generation of ATP by ACK. In most organisms tested, pta is located either immediately downstream of xsc or in the immediate vicinity (65, 387; for a review, see reference 94).
Phosphoketolase.
The phosphoketolase pathway (PKP) metabolizes pentose sugars and sugar alcohols and, in some organisms under aerobic conditions, also certain hexoses, including glucose and gluconate. Xylulose 5-phosphate phosphoketolase (XPK), the central enzyme of the PKP, converts xylulose 5′-phosphate (X5P) into glyceraldehyde 3-phosphate and acetyl∼P (Fig. 9C) (302). XPK has been identified in heterofermentative lactobacilli, Acetobacter xylinum, Thiobacillus novellus, Butyrivibrio fibrisolvens, Fibrobacter succinogenes, Fibrobacter intestinalis, and yeasts (350). Note that XPK is unsimilar to the transketolases (TKT) used by many species (including E. coli) for conversion of X5P. XPK utilizes inorganic phosphate instead of phosphorylated aldose, it produces acetyl∼P instead of fructose-6-phosphate (F6P), and its sequence only poorly resembles that of TKT (350).
Two variants of XPK exist in Bifidobacteria spp.: one specific for F6P and one that recognizes both X5P and F6P (XFP) (301). The xfp gene, which encodes the dual-substrate enzyme, resides just upstream of pta (301). This arrangement, reminiscent of xsc and pta, suggests that the acetyl∼P formed by XFP also may perform dual metabolic functions: anabolism and energy conservation (387). CcpA-dependent carbon catabolite repression inhibits xpkA expression in Lactobacillus pentosus (350), the opposite of its effect on PTA and ACK in B. subtilis (164, 353). Since the PTA-ACK pathway in B. subtilis operates in a strictly catabolic role, this regulatory arrangement suggests that XPK and its homolog XFP operate primarily in their anabolic role.
Glycine reductase.
Some strictly anaerobic gram-positive amino-acid-degrading bacteria, including Clostridium sticklandii (422), Eubacterium acidaminophilum (401), and Tissierella creatinophila (179), form acetyl∼P during the reductive deamination of glycine, sarcosine, or betaine (16, 17, 199, 305, 423). The enzymes responsible, e.g., glycine reductase, consist of several subunits. The substrate specificity resides in substrate-activating proteins B (12), which convert their substrate to a common carboxymethyl residue attached to selenoprotein A while evolving ammonia, monomethylamine or trimethylamine. In the presence of inorganic phosphate, protein C oxidizes carboxymethyl selenoprotein A with the transfer of the acetyl group to phosphate to from acetyl∼P (462). Energy is conserved because ACK converts the acetyl∼P to ATP (Fig. 9D) (17; for reviews, see references 11 and 12).
“FLIPPING THE SWITCH”: REGULATING acs TRANSCRIPTION
E. coli cells switch from acetate dissimilation to acetate assimilation primarily by controlling the initiation of acs transcription (249), although other regulatory mechanisms may contribute (384, 426). This regulation appears quite complex. At minimum, it involves at least two promoters (40, 63, 249, 250), two sigma factors (250, 410), the transcription factor CRP (40, 249), and two nucleoid-associated proteins: IHF and FIS (62, 63). Published (249) and preliminary evidence (D. F. Browning, C. M. Beatty, S. Busby, and A. J. Wolfe, unpublished data) suggest that several other factors may participate.
Expression Profile
Growth on excess glucose represses AMP-ACS activity, while growth on acetate induces it (61). The same is true of acs transcription (249, 329). During growth on glucose in a buffered minimal medium, acs transcription remains low during acetogenic exponential growth (249, 329, 410, 451) (Fig. 10). Transcription induces during the transition from exponential growth to stationary phase, as cells coassimilate the remaining sugar and the previously excreted acetate (249, 410). Transcription peaks as the culture enters stationary phase and is followed by a rapid decrease. Transcription again rises throughout the first several hours of stationary phase, even though the concentration of acetate remains low (Wolfe and Beatty, unpublished). A similar, but not identical, pattern of expression ensues during growth in unbuffered tryptone broth. acs transcription induces as serine consumption concludes, aspartate consumption commences, and acetyl-CoA and acetyl∼P concentrations peak (62, 249, 360). acs transcription reaches its maximum as the culture enters stationary phase (249), decreases rapidly, and then slowly increases in parallel with extracellular acetate (Wolfe and Beatty, unpublished). Since acs transcription rises during early stationary phase whether or not extracellular acetate accumulates, one must look elsewhere for the cause of increased transcription during stationary phase.
FIG. 10.
Transcription of acs by cells grown in glucose minimal medium relative to optical density, consumption of carbon sources, excretion of acetate, and intracellular concentrations of FIS and IHF. OD, optical density.
acs Operon and Promoter Architecture
acs is the first gene in an operon that includes yjcH, a hypothetical gene, and actP (formerly known as yjcG), which encodes an acetate permease (153). No evidence exists for internal promoters; thus, transcription of the acs operon apparently initiates only from the region 5′ of the acs open reading frame (Fig. 11A). acs transcription occurs from two promoters; the proximal acsP2 functions as the primary acs promoter (40), while the distal acsP1 is a weak, secondary promoter located about 200 bp upstream of acsP2 (40, 63, 249, 250). acsP1 overlaps extensively with pnrfA, a divergent promoter (63), which drives the expression of a nitrite reductase (111).
FIG. 11.
Regulation of acs transcription. (A) Organization of the nrf-acs locus. The bent arrows represent transcription initiation sites. (B) nrf-acs intergenic region, showing the location of binding sites for CRP, FIS, and IHF. The numbers are relative to the transcription initiation site of acsP2. Numbers in parentheses are relative to the transcription initiation site of acsP1. (C) Proposed interactions for synergistic class III CRP-dependent activation. Cross-hatched double oval, CRP dimer; light gray ovals, α-CTD. Although RNAP possesses only two α-CTDs, several are depicted to demonstrate vacillation amongst various possible sites. Other gray and cross-hatched shapes represent the rest of RNAP.
Independent Regulation of Overlapping Promoters
The acs gene and the nrf operon encode products that perform vastly different functions under widely different conditions. acs induction permits cells to assimilate acetate (251), while nrfA induction occurs under anaerobic conditions, especially in the presence of nitrite (111). Despite their extensive overlap, pnrfA and acsP1 transcribe independently. Mutations that disrupt the −10 element of either promoter do not significantly influence initiation from the other (63). This independence probably arises from the fact that acsP1 initiates transcription infrequently. This repressed state appears to result from the combined action of the nucleoid proteins FIS and IHF, the oxygen regulator FNR, and the nitrite-responsive RR NarL (63) (see below).
σ70-Dependent Transcription
To seek the sigma factor most responsible for acs expression, Kumari et al. used isogenic strains that carry a temperature-sensitive allele of rpoD (which encodes the housekeeping sigma factor σ70) and either a wild-type or null allele of rpoS (which encodes σS). On the basis of immunoblot analysis, reverse transcription-PCR, and acs::lacZ transcriptional fusion analyses performed on cells containing at most two copies of the acs promoter region, they concluded that acs induction during the transition from exponential growth to stationary phase depends on σ70. Cells that lacked a functional σ70 did not assimilate extracellular acetate, exhibited barely detectable levels of AMP-ACS, synthesized no detectable acs transcript, and displayed little acs promoter activity (250).
Some controversy exists concerning the role of σS. One study concluded that σS helps initiate acs transcription, because transcription was slightly reduced and delayed in rpoS mutants (410). A later study concluded that σS inhibits acs transcription after entry into stationary phase, because acs transcription did not decrease in rpoS mutants as it does in wild-type cells (250). Since the steady-state levels of σS increase during entry into stationary phase (183), the authors suggested that the rising pool of σS may inhibit acs transcription by competing with σ70 for core polymerase, as proposed previously (125). This competition would reduce acs transcription to levels appropriate for stationary phase (250). The discrepancy between these two studies may result from several factors, including the use of different genetic backgrounds, temperatures, and media. It also may result from the reliance on multicopy transcriptional fusions in the earlier study (410), whereas the later study involved diverse analyses of cells containing one or two copies of the acs promoter region (250).
Activation of acsP2 by CRP
In vitro, RNA polymerase (RNAP) alone does not transcribe acsP2 efficiently, although it binds and melts an extensive region of DNA. For efficient transcription, it requires CRP, apparently because CRP restricts open-complex formation to the region flanked by nucleotides +4 and −15, which includes the −10 element of acsP2. Thus, CRP facilitates transcription by “focusing” RNAP to acsP2 (40).
Upstream of acsP2, CRP binds to two sites (CRP I and CRP II), each located in class I positions (Fig. 11B). Each of these DNA sites permits a CRP dimer to recruit RNAP to the acsP2 promoter by contacting the C-terminal domain of each of the two α subunits of RNAP (α-CTD). An explanation of simple CRP-dependent activation including class I is given in Fig. 12. For a review, see reference 64. The higher-affinity site, CRP I (centered at position −69.5), is absolutely required for acsP2 transcription. The lower-affinity site, CRP II (centered at position −122.5), is necessary for maximal activation. CRP-dependent activation of acsP2 transcription involves activation region 1 (AR1) of CRP but does not involve AR2 (Fig. 11C). Activation also involves the 287, 261, and 265 determinants of the C-terminal domain of the α-CTD. Together, these observations are consistent with a synergistic class III mechanism by which tandem CRP proteins, each located at a class I position, activate acsP2 transcription by interacting with the α-CTD (40).
FIG. 12.
Simple CRP regulation operates through two related mechanisms, designated class I and class II. Both classes depend on specific interactions between CRP and RNAP. At class I promoters, a CRP dimer (cross-hatched double oval) binds to DNA at a site centered near position −61.5, −71.5, −82.5, or −92.5. CRP bound at any of these positions uses a defined surface (activating region 1 [AR1]) in the downstream subunit of CRP to contact a specific surface determinant (287) of the C-terminal domain of the α-subunit of RNAP (α-CTD; gray double oval). Two additional α-CTD determinants contribute to class I activation: the 261 determinant, proposed to interact with the σ subunit of RNAP (checkered shape), and the 265 determinant, which binds DNA, preferably A+T-rich sequences, e.g., the UP element. At class II promoters, a CRP dimer binds to DNA at a site centered near position −41.5. When bound at this position, CRP uses AR1 of the upstream subunit of CRP to contact determinant 287 of the α-CTD. CRP uses a second surface (AR2) of the downstream subunit to contact the 162-165 determinant of the N-terminal domain of α. The 265 determinant of the α-CTD also contributes by binding DNA. Class III promoters use more complex mechanisms that utilize two CRP dimers to achieve maximal transcription activation. In class III activation, the CRP dimers can bind either to tandem class I positions (as is the case at acs) or to one class II and one class I position. For reviews, see references 64, 69, and 158.
Negative Regulation by Nucleoid Proteins
Histone-like proteins, e.g., FIS and IHF, help compact the chromosome into a folded structure called the nucleoid (reviewed in references 21 and 345). These nucleoid-associated proteins also play dynamic and highly specialized roles, facilitating transient localized rearrangements of chromosome architecture that can influence transcription (reviewed in reference 299). Cells regulate FIS and IHF levels with respect to their physiological status (22, 23). FIS, the most abundant nucleoid-associated protein found after dilution of cells into fresh medium, diminishes in amount throughout exponential growth, becoming undetectable by stationary phase. In contrast, IHF levels increase steadily as the growth rate decreases to become, in stationary phase, the second most abundant nucleoid-associated protein. Thus, acs transcription remains low when FIS levels are high, peaks as FIS becomes undetectable, and then diminishes as IHF becomes abundant (Fig. 10).
Negative regulation by FIS.
FIS can negatively regulate CRP-dependent acsP2 transcription during early exponential phase by using both indirect and direct mechanisms. Since FIS represses crp transcription (155), it can preclude acs transcription by restricting the availability of CRP. Within the nrfA-acs intergenic region, FIS binds to several lower-affinity sites and three higher-affinity sites, FIS I, FIS II, and FIS III, centered at positions −267, −98, and −59, respectively, relative to the acsP2 transcription initiation site (Fig. 11B) (62). Thus, FIS also can regulate acs transcription directly, apparently using three distinct mechanisms. (i) FIS may repress acsP2 transcription by steric hindrance, since it can bind weakly to a DNA site that overlaps the −35 element of acsP2 (62). (ii) FIS can anti-activate CRP-dependent acsP2 transcription, since it clearly interferes with the binding of CRP. Note that FIS II (−98) lies between CRP II (−122.5) and CRP I (−69.5), while FIS III (−59) overlaps CRP I. Competitive DNase I footprint and electrophoretic mobility shift analyses and in vitro transcription assays indicate that FIS can displace CRP from both its sites. In vivo, a mutation in FIS II that diminishes its affinity for FIS by more than 10-fold increases acs transcription 2- to 3-fold during growth in tryptone broth. A similar increase results from a mutation that favors the binding of CRP over that of FIS to their overlapping sites (CRP I and FIS III). Thus, the competition between FIS and CRP for binding to their overlapping and tandem sites helps to keep acsP2 transcription low. (iii) Relative to acsP1, FIS I is centered at position −60 (Fig. 11B). In vivo, a fis mutant exhibits threefold more transcription from acsP1, suggesting that FIS helps keep this promoter repressed. The distal location of this site suggests a mechanism that does not involve steric hindrance of RNAP (63).
Negative regulation by IHF.
It appears likely that IHF contributes to reduced acs transcription. Within the nrfA-acs intergenic region, it binds to three higher-affinity sites: IHF I to IHF III, centered at positions −226, −180, and −153, respectively (Fig. 11B) (62). The distal-most site (IHF I) is centered at position −19 relative to the acsP1 transcription initiation site. In vivo, a mutant that lacks IHF displays threefold more transcription from acsP1, suggesting that IHF helps FIS keep this secondary promoter repressed. Given the location of this binding site, IHF probably represses acsP1 transcription by steric hindrance, i.e., by interfering directly with RNAP binding (63). In vitro transcription assays show that IHF decreases CRP-dependent acsP2 transcription by about twofold (62). The mechanism appears to involve a complex interaction between IHF, two CRP dimers, and RNAP. In vivo, the presence of IHF III strongly reduces CRP-dependent transcription, suggesting that the binding of IHF to this site interferes with the ability of CRP to activate transcription (62). This reduced transcription requires the integrity of IHF III, CRP II, the AR2 surface of CRP, and CRP I. Loss of the first three elements yields activity close to that of the wild-type promoter, while loss of the last element eliminates activity altogether. Together with the observation that IHF can bind simultaneously with CRP, these data suggest a working model in which the binding of IHF to IHF III alters the interaction of one CRP dimer with the other and/or with RNAP itself (C. M. Beatty, D. Thach, and A. J. Wolfe, unpublished data).
Independent regulation of acs transcription by FIS and IHF.
Competitive DNase I footprint analyses show that FIS and IHF can bind the acs regulatory region simultaneously with no apparent effect on each other. In vitro transcription studies indicate that FIS and IHF inhibit CRP-dependent acsP2 transcription independently. These observations, coupled with the knowledge that cells inversely regulate their FIS and IHF pools (Fig. 10), support the hypothesis that these nucleoid proteins regulate CRP-dependent acs transcription by mediating the formation of temporally and spatially distinct nucleoprotein complexes (62). Thus, acs is not simply a substrate-induced, catabolite-repressed gene but, rather, a prime example of complex regulatory circuitry in which dynamic nucleoprotein complexes ensure that transcription occurs appropriately.
Other Regulatory Factors
In light of the putative PDHC bypass formed by POXB and AMP-ACS, it is worthwhile noting that poxB mutants exhibit two- to threefold decreases in both acs transcription (249) and AMP-ACS activity. In contrast, cells that constitutively express POXB exhibit increased AMP-ACS activity. Although the mechanisms by which POXB exerts its influence remains unknown, these observations suggest that AMP-ACS may be primarily responsible for activation of the acetate produced by POXB (2).
Mutants deficient for the GB enzyme ICL, the GB repressor IclR, or the activator of iclR transcription (FadR) exhibit acs transcription profiles that resemble that displayed by poxB mutants. This suggests the influence of some as yet unidentified signal of carbon flux (249). Note that no evidence exists for a direct effect by either IclR or FadR: although in vitro DNA binding experiments have not been performed, sequence analysis reveals no sequence with similarity to the consensus for either transcription factor (Wolfe, unpublished).
Whereas Cra positively regulates the genes that encode the GB and gluconeogenesis (319, 366, 389, 439), there exists no evidence that this global regulator exerts a significant effect on acs transcription (Beatty and Wolfe, unpublished). Finally, no evidence supports a role for arp, a putative gene predicted to encode an ankyrin-like protein and mistakenly annotated as an ACS regulatory protein (Beatty and Wolfe, unpublished).
Signals
To what signals does acs transcription respond? Clearly, acs transcription responds to catabolite repression through cAMP and its receptor protein, CRP (249). However, it also appears to respond to other environmental factors, including low oxygen partial pressure via FNR (249), depleted media (410), and elevated temperature (Beatty and Wolfe, unpublished). There also exists evidence for an acetate-associated signal, as suggested by the similar expression profiles exhibited by cells that lack POXB, IclR, FadR, or ICL (249, 410). The identity of this signal, however, remains unknown. For various reasons, it appears unlikely that acetyl-CoA, acetyl∼P, or acetate operate directly on acs transcription (249). In contrast, some ratio of these or related central metabolites might play such a role. For example, starved cells possess a low acetyl-CoA/CoASH ratio (84). Since acs replenishes the acetyl-CoA pool, it seems reasonable for a low acetyl-CoA/CoASH ratio to signal the timing of acs induction. This might explain why acs transcription is induced in response to spent medium that contains no acetate (410) and why acs transcription increases once both glucose and acetate become depleted. The mechanism by which this ratio might be sensed is not understood; however, signaling roles for acetyl-CoA and other acyl-CoAs have been documented (99, 221, 245, 266).
POST-TRANSCRIPTIONAL CONTROL
Post-Transcriptional Control by the Csr System
Mutants that lack CsrA, the csrB binding protein, exhibit three- to fourfold less AMP-ACS activity than do their wild-type parents. Thus, the Csr system positively regulates AMP-ACS activity. Although it is clear that this regulation operates on some post-transcriptional step (384), the mechanism remains elusive (T. Romeo, personal communication).
Post-Translational Control by Acetylation-Deacetylation
The activity of AMP-ACS is controlled post-translationally by an acetylation-deacetylation system. A newly identified protein acetyltransferase acetylates AMP-ACS (425). In AMP-ACS, acetylation occurs on a lysine residue (K609) invariant in all members of the firefly luciferase superfamily (426). The acetylated enzyme (AMP-ACS-Ac) is inactive (426), because acetylation inhibits the adenylylation of acetate (169) while the thioester-forming step remains unaffected (169, 200, 426). In vitro, AMP-ACS also acetylates itself, also on K609 (30). This may represent the source of the low-intensity signal observed in cells that lack PAT (425). AMP-ACS-Ac becomes reactivated by CobB, a prokaryotic member of the Sir2 (sirtuin) family of NAD+-dependent deacetylases (426), which have been implicated in gene silencing, cell aging, and chromatin remodeling (for reviews, see references 162, 166, 180, and 311). Since AMP-ACS-Ac (and presumably PrpE-Ac) is inactive, the growth of S. enterica on acetate or propionate as the sole carbon and energy source also depends on the ability of the CobB sirtuin to deacetylate AMP-ACS-Ac (428, 449, 450). Best known for their ability to deacetylate histones, human and yeast sirtuins expressed heterologously in S. enterica cobB mutants restore growth on either acetate or propionate. This observation, coupled with the presence of sirutins in all three kingdoms, has led Escalante-Semerena and colleagues to propose a universal role for sirtuins, connecting central metabolism to transcription and, perhaps, to other cellular functions that involve acetyltransferases (428).
AMP-ACS-Dependent Acetylation and Chemotaxis
Not only does it acetylate CoASH, AMP, and itself, but AMP-ACS also reversibly acetylates the chemotaxis RR CheY (30). To do so, it can use either acetyl-CoA or acetate (in the presence of ATP) as its acetyl donor (31). In either case, the intermediate is acetyl-AMP (31). Like CheY-P, CheY-Ac can generate CW rotation of the flagellar motor both in vitro and in vivo (26, 31, 364, 474). CheY purified from cell lysates appears to be a mixture of differentially and multiply acetylated species, with nonacetylated CheY being the most abundant. In vitro acetylation mediated by AMP-ACS increases the acetylation level of CheY. All the acetylation sites are lysine residues (30). Two of these (K92 and K109) become acetylated (364), and K92 is involved in the ability of AMP-ACS to activate CheY (27, 364). However, the double-mutant CheY (K92R K109R) protein can be acetylated by AMP-ACS to 70% of the wild-type level, consistent with multiple sites of acetylation (30).
Unlike acetyl∼P-dependent phosphorylation of CheY, the physiological consequence of CheY-Ac can be seen in the presence of the cognate HK CheA. Cells that lack AMP-ACS exhibit reduced sensitivity to both attractants and repellents. The normal response to repellents, but not to attractants, requires residue K92 of CheY (27). Although the role of CheA-P has been defined clearly, the mechanistic function of CheY-Ac remains obscure. The kinetics of acetylation preclude a direct role in excitation, and a role in adaptation also seems unlikely (27). In contrast to phosphorylation, acetylation generates CW rotation without increasing the binding of CheY to the flagellar switch. Thus, CheY-Ac must act on a step following switch binding (364).
An intriguing connection between phosphorylation and acetylation apparently exists, at least in vitro. First, both phosphoryl donors of CheY, CheA-P and acetyl∼P, each bind AMP-ACS and strongly inhibit CheY acetylation. Second, CheZ, the enzyme that enhances CheY dephosphorylation, enhances CheY acetylation. Finally, the presence of AMP-ACS raises the levels of both CheA-P and CheY-P. On the basis of these observations, Barak and Eisenbach propose that acetylation of CheY acts as a tuning system that compensates for cell-to-cell variations in the concentrations of CheA and CheZ (29). If so, such compensation would operate as cells transit from exponential growth to stationary phase and thus from growth on sugars to consumption of acetate.
Several groups have suggested a connection between acetate metabolism and chemotaxis (26, 364, 474). Flagellar biosynthesis begins during mid-exponential growth as acetyl∼P levels begin to diminish (360). Motility (and chemotaxis) reaches its maximum as E. coli cells transition from exponential growth to stationary phase (9), when the extracellular concentration of acetate and acs transcription both peak (249, 360). Since AMP-ACS-mediated acetylation of CheY increases the probability of CW rotation, its absence should decrease such behavior. However, a triple mutant lacking the three known routes to CheY activation (i.e., CheA, the PTA-ACKA pathway, and AMP-ACS) rotates its flagella CW more often than does a double mutant that retains AMP-ACS (364). This surprising observation may reflect the ability of CobB to drain acetyl groups from CheY through AMP-ACS. If so, then a mutant lacking CheA and the PTA-ACKA pathway should exhibit more CW behavior in the absence of CobB than in its presence, and the effect should depend upon AMP-ACS. If so, then the ability of CobB to regulate AMP-ACS-dependent acetylation of CheY might explain phosphorylation-independent chemotactic behaviors (27, 28).
ACETATE SWITCH IN OTHER ORGANISMS
In many bacteria and archaea, including E. coli and S. enterica, dissimilation depends on the PTA-ACKA pathway while assimilation acts through AMP-ACS. In other bacteria, some archaea, and certain lower eukaryotes, dissimilation depends on an ADP-forming acetyl-CoA synthetase (ADP-ACS). In still others, assimilation occurs via the ACKA-PTA pathway. Phylogenic and crystallographic studies have demonstrated the ubiquity of AMP-ACS (169, 227), ADP-ACS (130, 391), PTA (147), and ACKA (70, 71, 488). Below are described the well-characterized PTA-ACKA/AMP-ACS “acetate switch” of the gram-positive bacterium B. subtilis, acetate assimilation by the PTA-ACK pathway of the industrially important gram-positive bacterium Corynebacterium glutamicum, an ADP-ACS/AMP-ACS “acetate switch” found in halophilic archaea, the “propionate switch” of S. enterica, and the “acetate switch” of mammals.
PTA-ACKA/AMP-ACS Acetate Switch of the Gram-Positive Bacterium B. subtilis
Like E. coli, the gram-positive bacterium Bacillus subtilis excretes metabolic intermediates during aerobic growth on excess carbohydrates, during fermentation on glucose supplemented with pyruvate or a mixture of amino acids, or during anaerobic respiration using nitrate as the electron acceptor (192, 317). These overflow metabolites include the organic acids acetate, pyruvate, d-lactate, and succinate. They also include uncharged compounds such as ethanol, acetoin, and small amounts of 2,3-butanediol (317, 418). The conversion of carbon from sugar to excreted organic acid can be quite high. For example, when grown on a lightly buffered complex medium supplemented with glucose, the related B. cereus (a close relative of B. anthracis) excretes about 95% of the carbon as pyruvate and acetate (178).
Unlike E. coli, B. subtilis produces no formate, apparently because the organism does not possess the gene that encodes PFL (252). Mutants lacking a functional PDHC cannot grow anaerobically and do not excrete fermentation products. Thus, B. subtilis produces most, if not all, of its acetate from pyruvate via the PDHC and the PTA-ACKA pathway (317). Note that the gram-positive pathogen Enterococcus faecalis also uses PDHC to metabolize pyruvate during anaerobiosis (414).
Like E. coli, B. subtilis excretes acetate during exponential growth. During late exponential growth, it diverts some of the pyruvate to an uncharged compound, either acetoin (under aerobic conditions) or 2,3-butanediol (during anaerobiosis). The excretion of these uncharged compounds instead of the organic acids pyruvate and acetate avoids overacidification of the environment (418). During stationary phase, when the glucose has become depleted, B. subtilis assimilates and activates acetate via AMP-ACS (163) and acetoin by means of the acetoin dehydrogenase system (204), which oxidizes acetoin to acetyl-CoA and acetaldehyde with the production of one NADH (365).
Like E. coli, the B. subtilis PTA-ACKA pathway converts acetyl-CoA to acetate via an acetyl∼P intermediate (164, 353, 407). PTA plays a general role during anaerobic energy metabolism (365) and contributes to aerobic growth (353, 407). As in E. coli, mutants that lack pta grow poorly during anaerobiosis, although they reach rates close to wild type in aerobic and complex nutrient environments (365, 407). Although pta mutants excrete reduced amounts of acetate, especially during aerobic exponential growth on glucose and anaerobiosis (353, 365), they still manage to excrete substantial amounts of acetate during stationary phase. Thus, an additional acetate excretion pathway must exist; however, it does not involve the acetoin excretion pathway (353). Mutants that lack ACKA grow on complex medium about as rapidly as their wild-type parents do, except in the presence of excess glucose (164). Since pta mutants do not exhibit this behavior, this growth defect may be caused by an accumulation of acetyl∼P (164, 353).
Like E. coli and S. enterica, many bacteria organize pta and ack as a single operon (224, 259, 412, 435, 466). In contrast, the B. subtilis pta (formerly ywfJ) and ackA genes are located at distant loci on the chromosome (353, 407). Many organisms use a similar arrangement, including Mycoplasma genitalium (139), Lactobacillus sanfranciscensis (243, 244), Bifidobacterium lactis (301), and organisms that possess xsc (387). To coordinate the transcription of these distant genes, B. subtilis primarily uses CcpA (164, 165, 184). This ubiquitous member (253) of the LacI-GalR family of transcription factors (185) binds to conserved cre (formerly amyO) sites in the promoter regions of its target genes (184, 207, 236, 468). CcpA acts as a negative regulator of secondary carbon utilization genes, e.g., acsA. CcpA also functions as a positive regulator of genes involved in the excretion of excess carbon, e.g., pta, ackA, and alsSD, which encodes the acetoin production pathway (164, 184, 206, 353, 374, 453). Although CcpA is expressed constitutively in B. subtilis (184, 307), its activity is controlled by the status of either HPr (ptsH) or its paralog Crh (crh). HPr and Crh are phosphorylated and dephosphorylated by HPrK/P, an ATP-dependent kinase/phosphatase encoded by ptsK (116, 145, 146, 222, 372, 373). Nutritional status determines the balance between the kinase and phosphatase activities of HprK/P. Growth on glucose and, hence, high concentrations of the early glycolytic intermediates d-fructose-1,6-diphosphate and d-fructose-2,6-diphosphate favor kinase activity, while inorganic phosphate, glyceraldehyde-6-phosphate, and acetyl∼P favor phosphatase activity (248, 470). Mutations that disrupt this signaling pathway eliminate glucose repression of acsA and glucose activation of pta and ackA (116, 117, 353, 407, 452).
During growth on complex media, transcription of both pta and ackA peaks during mid-exponential growth and diminishes during entry into the stationary phase (353, 363, 452). The way in which cells achieve basal transcription of pta and ackA remains a mystery; however, for ackA, the binding of CcpA to a single cre site seems important (452). The presence of glucose further activates the transcription of both pta and ackA (164, 353). This increased transcription depends on the ability of activated CcpA to bind to cre sites located at positions −55.5 (pta) and −56.5 (ackA) relative to their respective transcription initiation sites (353, 407, 452). The molecular mechanism by which CcpA activates transcription is not known; although its E. coli ortholog Cra (367, 389) activates transcription of ppsA, which encodes a gluconeogenic enzyme, from a site located at position −45.5 (319). At these three promoters, CcpA and Cra probably sit on the same face of the helix; thus, they probably activate transcription by a similar mechanism (452). Additional factors probably influence the transcription of both ackA and pta. Both promoters possess two sequence elements located just upstream of their respective cre site, and activation of ackA transcription depends on these elements. The strong conservation of these elements and their similar location relative to cre suggest that similar mechanisms activate ackA and pta transcription, thereby providing the coordination that E. coli obtains by operon structure (312).
Additional information exists concerning the regulation of the PTA-ACKA pathway. Various stresses regulate the steady-state levels of PTA, as monitored by two-dimensional gel electrophoresis (13). However, the activity of the pathway is unaffected by oxygen tension, nitrate, or nitrite, and pta transcription does not depend on the anaerobic sensing systems ResDE (a 2CST pathway), FNR (365), or ArfM (formerly YwiD) (289). Transcription of pta also does not depend on the sporulation regulators SpoOA or SpoOH (407). Finally, pta may regulate its own gene because a strain carrying the intact open reading frame yields about twice as much pta promoter activity as does a strain deleted for pta (353).
Unlike E. coli, B. subtilis does not utilize the PTA-ACKA pathway for growth on acetate; instead, it relies exclusively on AMP-ACS, encoded by acsA (163). During growth on mixtures of amino acids, acsA transcription exhibits a diauxic pattern of expression: it is induced during midexponential growth, plateaus for several hours, and then is induced again 3 h into stationary phase. This pattern of expression suggests a preference for certain amino acids (165), not unlike that observed with E. coli (359). The mechanism by which cells achieve this pattern of expression remains unknown, although it does not depend on CcpA (165). In contrast, the mechanism by which excess glucose represses acsA transcription requires activated CcpA and a cre site (O2) located at position +44.5 (163, 165, 485). The downstream location of its DNA site suggests that CcpA might act as a transcriptional roadblock; however, CcpA-dependent repression does require the transcription-repair coupling factor (Mfd) (485).
Twenty base pairs upstream of the acs promoter (pacs), another promoter is located. This promoter, pacu, drives the expression of the divergently transcribed acu operon, which influences the utilization of acetoin by some unknown mechanism (163). acu does not encode AoDH ES, which is encoded by the aco operon (204). A second cre site (O1) overlaps the pacu −35 element. Disruption of O1 eliminates glucose repression of acu transcription, most probably because CcpA can no longer hinder RNAP binding. Disruption of this CcpA-cre interaction has no effect on acsA transcription or on glucose repression of that transcription. Similarly, disruption of the other cre site, O2, has no effect on acu transcription. However, it not only eliminates glucose repression of acsA transcription but, surprisingly, also permits the activation of acsA transcription in response to glucose. Several possible mechanisms have been proposed for this behavior (165). Repression probably involves the binding of CcpA to both cre sites. In the absence of binding to the downstream site O2, CcpA can bind only O1. Since it is centered at position −63.5 relative to the transcription initiation site of pacsA, it might activate transcription by making contact with RNAP, perhaps via its α-CTD. Alternatively, the second factor may not be CcpA. Indeed, CodY, a GTP-sensing transcription factor, has been reported to repress acsA (131, 485).
Acetate Assimilation by the PTA-ACK Pathway of an Industrial Corynebacterium sp.
C. glutamicum, a gram-positive bacterium used widely for the production of amino acids (267), can grow on either glucose or acetate as its sole carbon and energy source. Unlike E. coli or B. subtilis, wild-type cells do not accumulate substantial amounts of acetate during aerobic growth on glucose (470), although they may excrete acetate during anaerobic growth (120). Like E. coli, C. glutamicum uses the PTA-ACK pathway to assimilate high concentrations of acetate (200 mM). Assimilation also requires the GB. Under these conditions, mutants that lack the PTA-ACK pathway or the GB do not grow (369, 470). Unlike E. coli and B. subtilis, this organism appears to have no catabolite repression system (152). Thus, wild-type C. glutamicum cometabolizes acetate (e.g., 100 mM) in the presence of high concentrations of glucose (e.g., 55 mM) (469, 470). However, PTA-ACK pathway mutants also cometabolize glucose and acetate (152). Thus, an alternative acetate assimilation pathway must exist, although AMP-ACS is an unlikely candidate: AMP-ACS activity has not been detected, and an obvious acs homolog does not appear in the genome sequence (152). Unlike E. coli, C. glutamicum requires the PTA-ACK pathway to grow on propionate (152) and no obvious prpE homolog appears in the genome (88). C. glutamicum appears to exert most of its control of the pta ack operon and the GB genes aceA and aceB primarily at the level of transcription initiation, which correlates with the presence of acetate in the environment (152, 314, 369, 469, 470). The transcription factor RamB binds sequences located upstream of the pta ack operon and within the aceA-aceB intergenic region. During growth on glucose, the binding of RamB represses transcription of all four genes. During growth on acetate, it contributes to full expression of aceA and aceB (151). For a review, see reference 152.
ADP-ACS/AMP-ACS Acetate Switch of Halophilic Archaea
Like E. coli, some halophilic archaea (Halococcus saccharolyticus, Haloferax volcanii, and Halorubrum saccharovorum) undergo an acetate switch during growth on glucose. Like E. coli, these organisms excrete acetate during exponential growth and cometabolize acetate and glucose as they enter stationary phase. Like E. coli, acetate assimilation requires an inducible AMP-ACS. Unlike E. coli, these acetogenic organisms do not possess the PTA-ACKA pathway. Instead, they dissimilate acetate by means of an ADP-forming acetyl-CoA synthetase (ADP-ACS [EC6.2.1.13]), an enzyme distinct from AMP-ACS and instead related to the SCSC (59). Like PTA and ACKA, ADP-ACS converts acetyl-CoA, inorganic phosphate, and ADP into acetate, CoASH, and ATP. Unlike PTA and ACKA, it performs this conversion in one step (400). Originally identified in the eukaryotic protists Entamoeba histolytica and Giardia lamblia (368, 392), ADP-forming ACS has been found in all acetate-forming archaea studied to date, including anaerobic hyperthermophiles and mesophilic aerobic halophiles (154, 285, 316, 395, 396).
“Propionate Switch” of S. enterica
Cells of S. enterica growing on 1,2-propanediol excrete propionate, presumably via a coenzyme B12-dependent pathway (331). Two pathways can activate propionate to propionyl-CoA. The first, catalyzed by either PrpE or AMP-ACS, involves a propionyl-AMP intermediate. Deletion of both enzymes or deletion of the sirtuin deacetylase CobB, which activates both enzymes, does not influence the amount of propionate excreted. The second, catalyzed by a propionate kinase (encoded by pduW) and PTA, involves a propionyl∼P intermediate (52, 331). Deletion of either PduW or PTA results in a 50% increase in propionate excreted into the environment. ACKA can compensate for PduW only if overexpressed (331). Note that both E. coli and S. enterica also possess the inducible propionate kinase, encoded by tdcD, required for decarboxylation of threonine (186).
Mammalian “Acetate Switch”
An “acetate switch” also operates in mammals (96), where acetate is generated through both exogenous and endogenous mechanisms. For example, bacterial fermentation in the colon generates large amounts of exogenous acetate, which is transported via the portal vein to the liver, where AMP-ACS activates it to acetyl-CoA (66, 361, 398). This scenario is particularly true in ruminant animals (300). In nonruminants, the liver oxidizes ingested ethanol first to acetaldehyde and then to acetate, which AMP-ACS then activates to acetyl-CoA (97). Endogenously, acetate is not generated through an acetyl∼P intermediate. Instead, it is generated from acetyl-CoA through the action of acetyl-CoA hydrolase, a ubiquitous cytosolic enzyme (97). This permits the lipophilic acetate to diffuse freely across membranes that separate subcellular compartments. In the nervous system, acetylcholinesterase degrades the neurotransmitter acetylcholine, producing acetate, which AMP-ACS reactivates to replenish acetylcholine stores (75, 430). In the nucleus, histone deacetylases generate acetate, which must be reactivated to acetyl-CoA for reutilization (247).
Like yeast (455, 487), mammals possess two isoforms of AMP-ACS (144, 281, 475, 480), one cytosolic and the other mitochondrial. The activity of the cytosolic enzyme (AceCS1), found predominantly in the liver, activates acetate to supply cells with acetyl-CoA for lipid synthesis (281, 475). AceCS1 activity responds to changes in nutritional and hormonal status (475). Transcription of AceCS1 is regulated by at least two members of the sterol regulatory element-binding protein (SREBP) family (144, 210, 281). Like Ino2p and Ino4p, proteins that regulate yeast acs transcription (189), SREBP family members are basic helix-loop-helix transcription factors that mediate cholesterol and fatty acid biosynthesis (202). AceCS1 transcripts accumulate in the livers of transgenic mice that overexpress SREBP and are almost absent in SREBP-null mice. Fasting reduces transcript levels, while refeeding restores them. Diabetic mice express low levels, which can be induced by insulin (416). In contrast, the mitochondrial enzyme (AceCS2) is found predominantly in heart and skeletal muscle but also in the spleen, lungs, kidneys, testes, and brain (144). Ketogenic conditions, e.g., starvation or diabetes, induce AceCS2 expression. Under such conditions, the liver releases substantial amounts of acetate, which accumulates in the bloodstream (66, 405, 481). AceCS2 activates this acetate, providing acetyl-CoA to the TCA cycle, where it is oxidized for the generation of ATP (144).
AMP-ACS also may be regulated developmentally. During mouse embryogenesis, for example, AceCS1 transcripts accumulate differentially in the developing nervous system and during organogenesis (276), a situation similar to that observed in developing Arabidopsis seeds, where it accumulates within chloroplasts (230).
CONCLUDING REMARKS
In this postgenomic era, we now possess the tools to discover how cells coordinate their central metabolic pathways—with each other, with those that lead to secondary metabolites, with essential cellular processes, with organelle biogenesis, and with the circuitry and machinery that make these processes possible. It goes without saying that this discovery process must pass through acetyl-CoA, the keystone of central metabolism.
The molecular switch that interconverts acetyl-CoA and acetate not only supplies the cell with opportunities to recover much needed NAD+, recycle limited stores of CoASH, and generate ATP, but it also produces compounds that can pass freely through membranes. For bacteria, the first half of this switch, acetate dissimilation, permits cells to grow rapidly when carbon is present in excess over oxygen or nitrogen, while the second half allows cells to reclaim that excreted acetate. For eukaryotes, this switch facilitates the passage of two-carbon units from one compartment to another, for example permitting the pyruvate formed by glycolysis (in the cytosol) to be fully oxidized by the TCA cycle (within the mitochondrial matrix).
Each half of the acetate switch furnishes the cell with a prime opportunity to monitor its nutritional status and the likelihood of its internal acidification and to use that information to coordinate its gene expression and cellular processes. Its instability and the tendency of its pool to rise and fall as a function of carbon, nitrogen, oxygen, and phosphate fluxes through central metabolic pathways make acetyl∼P an excellent global signal. Clearly, the mechanism(s) by which acetyl∼P exerts its influence must be clarified. However, its propensity in vitro to donate phosphoryl groups, especially to RRs of 2CST pathways, suggests an obvious mechanism—one which remains to be tested definitively. Of course, other mechanisms also must be considered seriously. In some cases, acetyl∼P may represent an alternative source of energy, as proposed for peroxide resistance in S. pneumoniae (343). Alternatively, it may function as a ligand for the activation of diverse enzymes, as it does in vitro for B. subtilis HprK/P (470) and ATP-PFK of Desulfurococcus amylolyticus (177). In all probability, acetyl∼P influences cellular processes through a combination of mechanisms.
The extent to which acetyl∼P influences gene expression and other cellular processes also remains to be determined. If acetyl∼P interacts with even a small subset of RRs, the extent is likely to be vast. The knowledge that acetyl∼P influences the activity of RRs in both gram-negative and gram-positive bacterium suggests a universal relationship. Since most bacteria possess 2CST systems, many of which regulate virulence factors, this relationship probably contributes to the pathogenicity of many organisms, including many potential biological weapons. Given that mammals do not synthesize acetyl∼P or possess 2CST pathways, the interface between acetyl∼P and 2CST pathways represents a prime target for pharmaceutical therapies or immune strategies.
The assimilation half of the “acetate switch” presents the cell with a second opportunity to monitor nutritional status. It makes perfect sense for the presence or absence of covalently bound acetyl groups to regulate AMP-ACS, an enzyme whose sole purpose is to activate acetate by acetylating AMP and then CoASH. It also makes sense for the acetylation state of that enzyme to depend upon an NAD+-dependent activity. It would be detrimental to cells that had not sufficiently regenerated NAD+ to begin assimilating acetate: MDH, a key enzyme in both the GB and the TCA cycle, requires NAD+. This NAD+-dependent regulation of AMP-ACS also suggests a mechanism for phosphoryl-independent chemotaxis. Through AMP-ACS, fluctuations in the intracellular NAD+ concentration could regulate CheY activity and thus the probability of CW flagellar motor rotation. Using such a mechanism, a swimming cell could sense the direction of a gradient of some assimilable carbon source, because its consumption alters the balance between NAD+ and NADH. Perhaps this represents the primordial chemotaxis machinery, which permitted motile cells to scavenge for food prior to the evolution of histidine kinases and chemoreceptors.
The complexity of acs transcription should not be surprising considering the importance of acetate assimilation. We expect that our dissection of the acs transcriptional machinery will teach us fundamental lessons about how bacterial cells regulate the vast number of similarly complex promoters. In particular, it will be fascinating to see how the component that recognizes the specific acetate-associated signal integrates into the overall, already quite complex, transcription scheme. On this note, one might consider the possibility that this missing signal may be related to the ratio of NAD+ to NADH or, perhaps, of acetyl-CoA to CoASH.
Finally, we must fully invest in efforts to understand the “acetate switch” of other organisms. Relative to E. coli and S. enterica serovar Typhimurium, we know almost nothing concerning acetate metabolism in other model organisms, clinically important pathogens, industrial production strains, and ourselves.
Acknowledgments
The work performed in the Wolfe laboratory has been supported by grants from the National Institutes of Health, the National Science Foundation, and Loyola University Chicago.
I thank all my collaborators, all the individuals who provided me with access to their unpublished results, and anyone with whom I have spent hours discussing various aspects of the “acetate switch.” I specifically want to emphasize the intellectual and technical efforts of four postdoctoral fellows, Christine M. Beatty, Douglas F. Browning, Suman Kumari, and Birgit M. Prüβ.
REFERENCES
- 1.Abaibou, H., J. Pommier, S. Benoit, G. Giordano, and M. Mandrand-Berthelot. 1995. Expression and characterization of the Escherichia coli fdo locus and a possible physiological role for aerobic formate dehydrogenase. J. Bacteriol. 177:7141-7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdel-Hamid, A. M., M. M. Attwood, and J. R. Guest. 2001. Pyruvate oxidase contributes to the aerobic growth efficiency of Escherichia coli. Microbiology 147:1483-1498. [DOI] [PubMed] [Google Scholar]
- 3.Alam, K. Y., and D. P. Clark. 1989. Anaerobic fermentation balance of Escherichia coli as observed by in vivo nuclear magnetic resonance spectroscopy. J. Bacteriol. 171:6213-6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexeeva, S., B. de Kort, G. Sawers, K. J. Hellingwerf, and M. J. de Mattos. 2000. Effects of limited aeration and of the ArcAB system on intermediary pyruvate catabolism in Escherichia coli. J. Bacteriol. 182:4934-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen, A. 1984. The structure and function of gastrointestinal mucus, p. 3-11. In I. E. C. Boedeker (ed.), Attachment of organisms to the gut mucosa, vol. II. CRC Press, Inc., Boca Raton, Fla. [Google Scholar]
- 6.Amarasingham, C. D., and B. D. Davis. 1965. Regulation of delta-ketoglutarate dehydrogenase formation in Escherichia coli. J. Biol. Chem. 240:3664-3668. [PubMed] [Google Scholar]
- 7.Amemura, M., K. Makino, H. Shinagawa, and A. Nakata. 1990. Cross talk to the phosphate regulon of Escherichia coli by PhoM protein: PhoM is a histidine protein kinase and catalyzes phosphorylation of PhoB and PhoM opon reading frame 2. J. Bacteriol. 172:6300-6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ames, G. F., and A. K. Joshi. 1990. Energy coupling in bacterial periplasmic permeases. J. Bacteriol. 172:4133-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amsler, C. D., M. Cho, and P. Matsumura. 1993. Multiple factors underlying the maximum motility of Escherichia coli as cultures enter post-exponential growth. J. Bacteriol. 175:6238-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson, G. G., J. J. Palermo, J. D. Schilling, R. Roth, J. Heuser, and S. J. Hultgren. 2003. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301:105-107. [DOI] [PubMed] [Google Scholar]
- 11.Andreesen, J. R. 1994. Glycine metabolism in anaerobes. Antonie Leeuwenhoek 66:223-237. [DOI] [PubMed] [Google Scholar]
- 12.Andreesen, J. R., M. Wagner, D. Sonntag, M. Kohlstock, C. Harms, T. Gursinsky, J. Jager, T. Parther, U. Kabisch, A. Grantzdorffer, A. Pich, and B. Sohling. 1999. Various functions of selenols and thiols in anaerobic gram-positive, amino acids-utilizing bacteria. Biofactors 10:263-270. [DOI] [PubMed] [Google Scholar]
- 13.Antelmann, H., J. Bernhardt, R. Schmid, H. Mach, U. Volker, and M. Hecker. 1997. First steps from a two-dimensional protein index towards a response-regulation map for Bacillus subtilis. Electrophoresis 18:1451-1463. [DOI] [PubMed] [Google Scholar]
- 14.Argenzio, R. A., and M. Southworth. 1974. Sites of organic acid production and absorption in gastrointestinal tract of the pig. Am. J. Physiol. Endocrinol. Metab. 228:454-460. [DOI] [PubMed] [Google Scholar]
- 15.Aristidou, A. A., K.-Y. San, and G. N. Bennett. 1994. Modification of central pathway in Escherichia coli to reduce acetate accumulation by heterologous expression of the Bacillus subtilis acetolactate synthase gene. Biotechnol. Bioeng. 44:944-951. [DOI] [PubMed] [Google Scholar]
- 16.Arkowitz, R., and R. Abeles. 1989. Identification of acetyl phosphate as the product of clostridial glycine reductase: evidence for an acetyl enzyme intermediate. Biochemistry 28:4639-4644. [DOI] [PubMed] [Google Scholar]
- 17.Arkowitz, R., and R. Abeles. 1991. Mechanism of action of clostridial glycine reductase: isolation and characterization of a covalent acetyl enzyme intermediate. Biochemistry 30:4090-4097. [DOI] [PubMed] [Google Scholar]
- 18.Arthur, M., F. Depardieu, G. Gerbaud, M. Galimand, R. Leclercq, and P. Courvalin. 1997. The VanS sensor negatively controls VanR-mediated transcriptional activation of glycopeptide resistance genes of Tn1546 and related elements in the absence of induction. J. Bacteriol. 179:97-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asada, Y., Miyake, M., Miyake, J., Kurane, R., and Tokiwa, Y. 1999. Photosynthetic accumulation of poly-(hydroxybutyrate) by cyanobacteria—the metabolism and potential for CO2 recycling. Int. J. Biol. Macromol. 25:37-42. [DOI] [PubMed] [Google Scholar]
- 20.Atkinson, M. R., and A. J. Ninfa. 1998. Role of the GlnK signal transduction protein in the regulation of nitrogen assimilation in Escherichia coli. Mol. Microbiol. 29:431-447. [DOI] [PubMed] [Google Scholar]
- 21.Azam, T. A., and A. Ishihama. 1999. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J. Biol. Chem. 274:33105-33113. [DOI] [PubMed] [Google Scholar]
- 22.Azam, T. A., A. Iwata, A. Nishimura, S. Ueda, and A. Ishihama. 1999. Growth phase- dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 181:6361-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ball, C. A., R. Osuna, K. C. Ferguson, and R. C. Johnson. 1992. Dramatic changes in FIS levels upon nutrient upshift in Escherichia coli. J. Bacteriol. 174:8043-8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bang, I., B. Kim, J. Foster, and Y. Park. 2000. OmpR regulates the stationary-phase acid tolerance response of Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:2245-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bang, I. S., J. P. Audia, Y. K. Park, and J. W. Foster. 2002. Autoinduction of the ompR response regulator by acid shock and control of the Salmonella enterica acid tolerance response. Mol. Microbiol. 44:1235-1250. [DOI] [PubMed] [Google Scholar]
- 26.Barak, R., W. N. Abouhamad, and M. Eisenbach. 1998. Both acetate kinase and acetyl coenzyme A synthetase are involved in acetate-stimulated chnage in the direction of flagellar rotation in Escherichia coli. J. Bacteriol. 180:985-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barak, R., and M. Eisenbach. 2001. Acetylation of the response regulator, CheY, is involved in bacterial chemotaxis. Mol. Microbiol. 40:731-743. [DOI] [PubMed] [Google Scholar]
- 28.Barak, R., and M. Eisenbach. 1999. Chemotactic-like response of Escherichia coli cells lacking the known chemotaxis machinery but containing overexpressed CheY. Mol. Microbiol. 31:1125-1137. [DOI] [PubMed] [Google Scholar]
- 29.Barak, R., and M. Eisenbach. 2004. Co-regulation of acetylation and phosphorylation of CheY, a response regulator in chemotaxis of Escherichia coli. J. Mol. Biol. 342:375-381. [DOI] [PubMed] [Google Scholar]
- 30.Barak, R., K. Prasad, A. Shainskaya, A. J. Wolfe, and M. Eisenbach. 2004. Acetylation of the chemotaxis response regulator CheY by acetyl-CoA synthetase purified from Escherichia coli. J. Mol. Biol. 342:383-401. [DOI] [PubMed] [Google Scholar]
- 31.Barak, R., M. Welch, A. Yanovsky, K. Oosawa, and M. Eisenbach. 1992. Acetyladenylate or its derivative acetylates the chemotaxis protein CheY in vitro and increases its activity at the flagellar switch. Biochemistry 31:10099-10107. [DOI] [PubMed] [Google Scholar]
- 32.Barker, H. A. 1992. The path from acetylphosphate to acetyl CoA. FASEB J. 6:3014-3015. [DOI] [PubMed] [Google Scholar]
- 33.Barnard, A., A. Wolfe, and S. Busby. 2004. Regulation at complex bacterial promoters: how bacteria use different promoter organisations to produce different regulatory outcomes. Curr. Opin. Microbiol. 7:102-108. [DOI] [PubMed] [Google Scholar]
- 34.Baronofsky, J. J., W. J. A. Schreurs, and E. R. Kashket. 1984. Uncoupling by acetic acid limits growth of and acetogenesis by Clostridium thermoaceticum. Appl. Environ. Microbiol. 48:1134-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baskett, R. C., and D. J. Hentges. 1973. Shigella flexneri inhibition by acetic acid. Infect. Immun. 8:91-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Basson, M. D., N. J. Emenaker, and F. Hong. 1998. Differential modulation of human (Caco-2) colon cancer cell line phenotype by short chain fatty acids. Proc. Soc. Exp. Biol. Med. 217:476-483. [DOI] [PubMed] [Google Scholar]
- 37.Batt, R. M., H. C. Rutgers, and A. A. Sancak. 1996. Enteric bacteria: friend or foe? J. Small Anim. Pract. 37:261-267. [DOI] [PubMed] [Google Scholar]
- 38.Bauer, D. A., A. Ben-Basst, M. Dawson, V. T. Dela Peunte, and J. O. Neway. 1990. Improved expression of human interleukin-2 in high-cell-density fermentor cultures of Escherichia coli K-12 by a phosphotransacetylase mutant. Appl. Environ. Microbiol. 56:1296-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bearson, B. L., L. Wilson, and J. W. Foster. 1998. A low pH-inducible, PhoPQ-dependent acid tolerance response protects Salmonella typhimurium against inorganic acid stress. J. Bacteriol. 180:2409-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beatty, C. M., D. F. Browning, S. J. W. Busby, and A. J. Wolfe. 2003. CRP-dependent activation of the Escherichia coli acsP2 promoter by a synergistic class III mechanism. J. Bacteriol. 185:5148-5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Begley, T. P., C. Kinsland, and E. Strauss. 2001. The biosynthesis of coenzyme A in bacteria. Vitam. Horm. 61:157-171. [DOI] [PubMed] [Google Scholar]
- 42.Behrens, M., and P. Durre. 2000. KdpE of Clostridium acetobutylicum is a highly specific response regulator controlling only the expression of the kdp operon. J. Mol. Microbiol. Biotechnol. 2:45-52. [PubMed] [Google Scholar]
- 43.Belaich, A., and J. P. Belaich. 1976. Microcalorimetric study of the anaerobic growth of Escherichia coli: growth thermograms in a synthetic medium. J. Bacteriol. 125:14-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bennett, P. M., and W. H. Holms. 1975. Reversible inactivation of the isocitrate dehydrogenase of Escherichia coli ML308 during growth on acetate. J. Gen. Microbiol. 87:37-51. [DOI] [PubMed] [Google Scholar]
- 45.Bentley, R. 2000. From ′reactive C2 units' to acetyl coenzyme A: a long trail with an acetyl phosphate detour. Trends Biochem. Sci. 25:302-305. [DOI] [PubMed] [Google Scholar]
- 46.Berg, P. 1956. Acyl adenylates: an enzymatic mechanism of acetate activation. J. Biol. Chem. 222:991-1013. [PubMed] [Google Scholar]
- 47.Bernish, B., and I. van de Rijn. 1999. Characterization of a two-component system in Streptococcus pyogenes which is involved in regulation of hyaluronic acid production. J. Biol. Chem. 274:4786-4793. [DOI] [PubMed] [Google Scholar]
- 48.Bertagnolli, B. L., and L. P. Hager. 1991. Activation of Escherichia coli pyruvate oxidase enhances the oxidation of hydroxyethylthiamin pyrophosphate. J. Biol. Chem. 266:10168-10173. [PubMed] [Google Scholar]
- 49.Bertagnolli, B. L., and L. P. Hager. 1993. Role of flavin in acetoin production by two bacterial pyruvate oxidases. Arch. Biochem. Biophys. 300:364-371. [DOI] [PubMed] [Google Scholar]
- 50.Blankenhorn, D., J. Phillips, and J. L. Slonczewski. 1999. Acid- and base-induced proteins during aerobic and anaerobic growth of Escherichia coli revealed by two- dimensional gel electrophoresis. J. Bacteriol. 181:2209-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bleves, S., M.-N. Marenne, G. Detry, and G. R. Cornelis. 2002. Up-regulation of the Yersinia enterocolitica yop regulon by deletion of the flagellum master operon flhDC. J. Bacteriol. 184:3214-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bobik, T. A., G. D. Havemann, R. J. Busch, D. S. Williams, and H. C. Aldrich. 1999. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. J. Bacteriol. 181:5967-5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bochner, B. R., and B. N. Ames. 1982. Selective precipitation orthophosphate from mixtures containing labile phosphorylated metabolites. Anal. Biochem. 122:100-107. [DOI] [PubMed] [Google Scholar]
- 54.Bock, A., and G. Sawers. 1996. Fermentation, p. 262-282. In F. C. Neidhardt, R. Curtiss, III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 55.Bohnhoff, M., C. P. Miller, and W. R. Martin. 1964. Resistance of the mouse's intestinal tract to experimental Salmonella infection. I. Factors which interfere with the initiation of infection by oral inoculation. J. Exp. Med. 120:805-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Booth, I. R. 1985. Regulation of cytoplasmic pH in bacteria. Microbiol. Rev. 49:359-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bouche, S., E. Klauck, D. Fischer, M. Lucassen, K. Jung, and R. Hengge-Aronis. 1998. Regulation of RssB-dependent proteolysis in Escherichia coli: a role for acetyl phosphate in a response regulator-controlled process. Mol. Microbiol. 27:787-795. [DOI] [PubMed] [Google Scholar]
- 58.Boucher, P. E., F. D. Menozzi, and C. Locht. 1994. The modular architecture of bacterial response regulators. Insights into the activation mechanism of the BvgA transactivator of Bordetella pertussis. J. Mol. Biol. 241:363-377. [DOI] [PubMed] [Google Scholar]
- 59.Brasen, C., and P. Schonheit. 2001. Mechanisms of acetate formation and acetate activation in halophilic archaea. Arch. Microbiol. 175:360-368. [DOI] [PubMed] [Google Scholar]
- 60.Brillard, J., C. Ribeiro, N. Boemare, M. Brehelin, and A. Givaudan. 2001. Two distinct hemolytic activities in Xenorhabdus nematophila are active against immunocompetent insect cells. Appl. Environ. Microbiol. 67:2515-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brown, T. D. K., M. C. Jones-Mortimer, and H. L. Kornberg. 1977. The enzymic interconversion of acetate and acetyl-coenzyme A in Escherichia coli. J. Gen. Microbiol. 102:327-336. [DOI] [PubMed] [Google Scholar]
- 62.Browning, D. F., C. M. Beatty, E. A. Sanstad, K. A. Gunn, S. J. W. Busby, and A. J. Wolfe. 2004. Modulation of CRP-dependent transcription at the Eschericheria coli acsP2 promoter by a nucleoprotein complex: anti-activation by the nucleoid proteins FIS and IHF. Mol Microbiol. 51:241-254 [DOI] [PubMed] [Google Scholar]
- 63.Browning, D. F., C. M. Beatty, A. J. Wolfe, J. A. Cole, and S. J. W. Busby. 2002. Independent regulation of the divergent Escherichia coli nrfA and acsP1 promoters by a nucleoprotein assembly at a shared regulatory region. Mol. Microbiol. 43:687-701. [DOI] [PubMed] [Google Scholar]
- 64.Browning, D. F., and S. J. W. Busby. 2004. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2:1-5. [DOI] [PubMed] [Google Scholar]
- 65.Bruggemann, C., K. Denger, A. M. Cook, and J. Ruff. 2004. Enzymes and genes of taurine and isethionate dissimilation in Paracoccus denitrificans. Microbiology 150:805-816. [DOI] [PubMed] [Google Scholar]
- 66.Buckley, B. M., and D. H. Williamson. 1977. Origins of blood acetate in the rat. Biochem. J. 166:539-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bulter, T., S. G. Lee, W. W. Wong, E. Fung, M. R. Connor, and J. C. Liao. 2004. Design of artificial cell-cell communication using gene and metabolic networks. Proc. Natl. Acad. Sci. USA 101:2299-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bunch, P. K., F. Mat-Jan, N. Lee, and D. P. Clark. 1997. The ldhA gene encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology 143:187-195. [DOI] [PubMed] [Google Scholar]
- 69.Busby, S., and R. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 70.Buss, K. A., D. R. Cooper, C. Ingram-Smith, J. G. Ferry, D. A. Sanders, and M. S. Hasson. 2001. Urkinase: structure of acetate kinase, a member of the ASKHA superfamily of phosphotransferases. J. Bacteriol. 183:680-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buss, K. A., C. Ingram-Smith, J. G. Ferry, D. A. Sanders, and M. S. Hasson. 1997. Crystallization of acetate kinase from Methanosarcina thermophila and prediction of its fold. Protein Sci. 6:2659-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Butow, R. A., and N. G. Avadhani. 2004. Mitochondrial signaling: the retrograde response. Mol. Cell 14:1-15. [DOI] [PubMed] [Google Scholar]
- 73.Cai, S. J., and M. Inouye. 2002. EnvZ-OmpR interaction and osmoregulation in Escherichia coli. J. Biol. Chem. 277:24155-24161. [DOI] [PubMed] [Google Scholar]
- 74.Carmany, D. O., K. Hollingsworth, and W. R. McCleary. 2003. Genetic and biochemical studies of phosphatase activity of PhoR. J. Bacteriol. 185:1112-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Carroll, P. T. 1997. Evidence to suggest that extracellular acetate is accumulated by rat hippocampal cholinergic nerve terminals for acetylcholine formation and release. Brain Res. 753:47-55. [DOI] [PubMed] [Google Scholar]
- 76.Chamnongpol, S., M. Cromie, and E. A. Groisman. 2003. Mg2+ sensing by the Mg2+ sensor PhoQ of Salmonella enterica. J. Mol. Biol. 325:795-807. [DOI] [PubMed] [Google Scholar]
- 77.Chamnongpol, S., and E. A. Groisman. 2000. Acetyl phosphate-dependent activation of a mutant PhoP response regulator that functions independently of its cognate sensor kinase. J. Mol. Biol. 300:291-305. [DOI] [PubMed] [Google Scholar]
- 78.Chang, D.-E., S. Shin, J.-S. Rhee, and J.-G. Pan. 1999. Acetate metabolism in a pta mutant of Escherichia coli W3110: importance of maintaining acetyl-CoA flux for the growth and survival. J. Bacteriol. 181:6656-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chang, Y. Y., and J. E. J. Cronan. 1983. Genetic and biochemical analyses of Escherichia coli strains having a mutation in the structural gene (poxB) for pyruvate oxidase. J. Bacteriol. 154:756-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chang, Y. Y., A. Y. Wang, and J. Cronan, J. E. 1994. Expression of Escherichia coli pyruvate oxidase (PoxB) depends on the sigma factor encoded by the rpoS (katF)gene. Mol. Microbiol. 11:1019-1028. [DOI] [PubMed] [Google Scholar]
- 81.Chatterjee, R., C. S. Millard, K. Champion, D. P. Clark, and M. I. Donnelly. 2001. Mutation of the ptsG gene results in increased production of succinate in fermentation of glucose by Escherichia coli. Appl. Environ. Microbiol. 67:148-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen, R., V. Hatzimanikatis, W. M. Yap, P. W. Postma, and J. E. Bailey. 1997. Metabolic consequences of phosphotransferase (PTS) mutation in a phenylalanine- producing recombinant Escherichia coli. Biotechnol. Prog. 13:768-775. [DOI] [PubMed] [Google Scholar]
- 83.Chiang, S. L., and J. J. Mekalanos. 1998. Use of signature-tagged transposon mutagenesis to identify Vibrio cholerae genes critical for colonization. Mol. Microbiol. 27:797-805. [DOI] [PubMed] [Google Scholar]
- 84.Chohnan, S., H. Furukawa, T. Fujio, H. Nishihara, and Y. Takamura. 1997. Changes in the size and composition of intracellular pools of nonesterified coenzyme A and coenzyme A thioesters in aerobic and facultatively anaerobic bacteria. Appl. Environ. Microbiol. 63:553-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chohnan, S., H. Izawa, H. Nishihara, and Y. Takamura. 1998. Changes in size of intracellular pools of coenzyme A and its thioesters in Escherichia coli K-12 cells to various carbon sources and stresses. Biosci. Biotechnol. Biochem. 62:1122-1128. [DOI] [PubMed] [Google Scholar]
- 86.Chohnan, S., and Y. Takamura. 1991. A simple micromethod for measurement of CoASH and its use in measuring intracellular levels of CoASH and short chain acyl- CoAs in Escherichia coli K12 cells. Agric. Biol. Chem. 55:87-94. [Google Scholar]
- 87.Chou, T. C., and F. Lipmann. 1952. Separation of acetyl transfer enzymes in pigeon liver extract. J. Biol. Chem. 196:89-103. [PubMed] [Google Scholar]
- 88.Claes, W. A., A. Puhler, and J. Kalinowski. 2002. Identification of two prpdbc gene clusters in Corynebacterium glutamicum and their involvement in propionate degradation via the 2-methylcitrate cycle. J. Bacteriol. 184:2728-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Clark, D. P. 1989. The fermentation pathways of Escherichia coli. FEMS Microbiol. Rev. 5:223-234. [DOI] [PubMed] [Google Scholar]
- 90.Clark, D. P., and J. E. Cronan, Jr. 1996. Two-carbon compounds and fatty acids as carbon sources, p. 343-357. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 91.Clegg, S., and K. T. Hughes. 2002. FimZ is a molecular link between sticking and swimming in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:1209-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Comolli, J. C., A. J., Carl, C. Hall, and T. Donohue. 2002. Transcriptional activation of the Rhodobacter sphaeroides cytochrome c2 gene promoter by the response regulator PrrA. J. Bacteriol. 184:390-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Contiero, J., C. M. Beatty, S. Kumari, C. L. DeSanti, W. R. Strohl, and A. J. Wolfe. 2000. Effects of mutations in acetate metabolism on high-cell-density growth of Escherichia coli. J. Ind. Microbiol. Biotechnol. 24:421-430. [Google Scholar]
- 94.Cook, A. M., and K. Denger. 2002. Dissimilation of the C2 sulfonates. Arch. Microbiol. 179:1-6. [DOI] [PubMed] [Google Scholar]
- 95.Cozzone, A. J. 1998. Regulation of acetate metabolism by protein phosphorylation in enteric bacteria. Annu. Rev. Microbiol. 52:127-164. [DOI] [PubMed] [Google Scholar]
- 96.Crabtree, B., M. J. Gordon, and S. L. Christie. 1990. Measurement of the rates of acetyl-CoA hydrolysis and synthesis from acetate in rat hepatocytes and the role of these fluxes in substrate cycling. Biochem. J. 270:219-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Crabtree, B., M. J. Souter, and S. E. Anderson. 1989. Evidence that the production of acetate in rat hepatocytes is a predominantly cytoplasmic process. Biochem. J. 257:673-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Crabtree, H. G. 1929. Observations on the carbohydrate metabolism of tumours. Biochem. J. 23:536-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cronan, J., Jr. 1997. In vivo evidence that acyl coenzyme A regulates DNA binding by the Escherichia coli FadR global transcription factor. J. Bacteriol. 179:1819-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cronan, J. E., Jr., and D. LaPorte. 1996. Tricarboxylic acid cycle and gloxylate bypass, p. 206-216. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 101.Cummings, J. H., and G. T. Macfarlane. 1997. Role of intestinal bacteria in nutrient metabolism. J. Parenter Enteral. Nutr. 21:357-365. [DOI] [PubMed] [Google Scholar]
- 102.Cummings, J. H., E. W. Pomare, W. J. Branch, C. P. Naylor, and G. T. Macfarlane. 1987. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28:1221-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cunning, C., and T. Elliott. 1999. RpoS synthesis is growth rate regulated in Salmonella typhimurium, but its turnover is not dependent on acetyl phosphate synthesis or PTS function. J Bacteriol. 181:4853-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cunningham, L., D. Georgellis, J. Green, and J. R. Guest. 1998. Co-regulation of lipoamide dehydrogenase and 2-oxoglutarate dehydrogenase synthesis in Escherichia coli: characterisation of an ArcA binding site in the lpd promoter. FEMS Microbiol. Lett. 169:403-408. [DOI] [PubMed] [Google Scholar]
- 105.Cunningham, L., and J. R. Guest. 1998. Transcription and transcript processing in the sdhCDAB-sucABCD operon of Escherichia coli. Microbiology 144:2113-2123. [DOI] [PubMed] [Google Scholar]
- 106.Dailey, F. E., and H. C. Berg. 1993. Change in direction of flagellar rotation in Escherichia coli mediated by acetate kinase. J. Bacteriol. 175:3236-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Danese, P. N., and T. J. Silhavy. 1998. CpxP, a stress-combative member of the Cpx regulon. J. Bacteriol. 180:831-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Danese, P. N., Snyder, W. B., Cosma, C. L., Davis, L. J. & Silhavy, T. J. 1995. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 9:387-398. [DOI] [PubMed] [Google Scholar]
- 109.Da Re, S. S., D. Deville-Bonne, T. Tolstykh, M. Veron, and J. B. Stock. 1999. Kinetics of CheY phosphorylation by small molecule phosphodonors. FEBS Lett. 457:323-326. [DOI] [PubMed] [Google Scholar]
- 110.Dartigalongue, C., and S. Raina. 1998. A new heat-shock gene, ppiD, encodes a peptidyl-prolyl isomerase required for folding of outer membrane proteins in Escherichia coli. EMBO J. 17:3968-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Darwin, A., H. Hussain, L. Griffiths, J. Grove, Y. Sambongi, S. Busby, and J. Cole. 1993. Regulation and sequence of the structural gene for cytochrome c552 from Escherichia coli: not a hexahaem but a 50 kDa tetrahaem nitrite reductase. Mol. Microbiol. 9:1255-1265. [DOI] [PubMed] [Google Scholar]
- 112.Davalos-Garcia, M., A. Conter, I. Toesca, C. Gutierrez, and K. Cam. 2001. Regulation of osmC gene expression by the two-component system rcsB-rcsC in Escherichia coli. J. Bacteriol. 183:5870-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.de Graef, M. R., S. Alexeeva, J. L. Snoep, and M. J. Teixeira de Mattos. 1999. The steady-state internal redox state (NADH/NAD) reflects the external redox state and is correlated with catabolic adaptation in Escherichia coli. J. Bacteriol. 181:2351-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Denger, K., J. Ruff, D. Schleheck, and A. M. Cook. 2004. Rhodococcus opacus expresses the xsc gene to utilize taurine as a carbon source or as a nitrogen source but not as a sulfur source. Microbiology 150:1859-1867. [DOI] [PubMed] [Google Scholar]
- 115.Deretic, V., J. H. J. Leveau, C. D. Mohr, and N. S. Hibler. 1992. In vitro phosphorylation of AlgR, a regulator of mucoidy in Pseudomonas aeruginosa, by a histidine protein kinase and effects of small phospho-donor molecules. Mol. Microbiol. 6:2761-2767. [DOI] [PubMed] [Google Scholar]
- 116.Deutscher, J., E. Kuster, U. Bergstedt, U. Charrier, and W. Hillen. 1995. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in Gram-positive bacteria. Mol. Microbiol. 15:1049-1053. [DOI] [PubMed] [Google Scholar]
- 117.Deutscher, J., J. Reizer, C. Fischer, A. Galinier, M. H. Saier, Jr., and M. Steinmetz. 1994. Loss of protein kinase-catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, by mutation of the ptsHI gene confers catabolite repression resistance to several catabolic genes of Bacillus subtilis. J. Bacteriol. 176:3336-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Diaz-Ricci, J. C., L. Regan, and J. E. Bailey. 1991. Effect of alteration of the acetic acid synthesis pathway on the fermentation pattern of Escherichia coli. Biotechnol. Bioeng. 38:1318-1324. [DOI] [PubMed] [Google Scholar]
- 119.Doelle, H. W., K. N. Ewings, and N. W. Hollywood. 1982. Regulation of glucose metabolism in bacterial systems. Adv. Biochem. Eng. 23:1-35. [Google Scholar]
- 120.Dominguez, H., C. Nezondet, N. D. Lindley, and M. Cocaign. 1993. Modified carbon flux during oxygen limited growth of Corynebacterium glutamicum and the consequences for amino acid overproduction. Biotechnol. Lett. 15:449-454. [Google Scholar]
- 121.Drake, S., R. Bourret, L. Luck, M. Simon, and J. Falke. 1993. Activation of the phosphosignaling protein Che Y. I. Analysis of the phosphorylated conformation by 19F NMR and protein engineering. J. Biol. Chem. 268:13081-13088. [PMC free article] [PubMed] [Google Scholar]
- 122.Durant, J. A., V. K. Lowry, D. J. Nisbet, L. H. Stanker, D. E. Corrier, and S. C. Ricke. 1999. Short-chain fatty acids affect cell-association and invasion of HEp-2 cells by Salmonella typhimurium. J. Environ. Sci. Health Ser. B 34:1083-1099. [DOI] [PubMed] [Google Scholar]
- 123.el-Mansi, E. M., and W. H. Holms. 1989. Control of carbon flux to acetate excretion during growth of Escherichia coli in batch and continuous cultures. J. Gen. Microbiol. 135:2875-2883. [DOI] [PubMed] [Google Scholar]
- 124.El-Mansi, M. 2004. Flux to acetate and lactate excretions in industrial fermentations: physiological and biochemical implications. J. Ind. Microbiol. Biotechnol. 31:295-300. [DOI] [PubMed] [Google Scholar]
- 125.Farewell, A., K. Kvint, and T. Nystrom. 1998. Negative regulation by RpoS: a case of sigma factor competition. Mol. Microbiol. 29:1039-1051. [DOI] [PubMed] [Google Scholar]
- 126.Farmer, W. R., and J. C. Liao. 1997. Reduction of aerobic acetate production by Escherichia coli. Appl. Environ. Microbiol. 63:3205-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Feng, J., M. R. Atkinson, W. McCleary, J. B. Stock, B. L. Wanner, and A. J. Ninfa. 1992. Role of phosphorylated metabolic intermediates in the regulation of glutamine synthetase synthesis in Escherichia coli. J. Bacteriol. 174:6061-6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ferre, A., J. de la Mora, T. Ballado, L. Camarena, and G. Dreyfus. 2004. Biochemical study of multiple chey response regulators of the chemotactic pathway of Rhodobacter sphaeroides. J. Bacteriol. 186:5172-5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ferrieres, L., and D. J. Clarke. 2003. The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface. Mol. Microbiol. 50:1665-1682. [DOI] [PubMed] [Google Scholar]
- 130.Field, J., B. Rosenthal, and J. Samuelson. 2000. Early lateral transfer of genes encoding malic enzyme, acetyl-CoA synthetase and alcohol dehydrogenases from anaerobic prokaryotes to Entamoeba histolytica. Mol. Microbiol. 38:446-455. [DOI] [PubMed] [Google Scholar]
- 131.Fisher, S. H. 1999. Regulation of nitrogen metabolism in Bacillus subtilis: vive la difference! Mol. Microbiol. 32:223-232. [DOI] [PubMed] [Google Scholar]
- 132.Forst, S., and B. Boylan. 2002. Characterization of the pleiotropic phenotype of an ompR strain of Xenorhabdus nematophila. Antonie Leeuwenhoek 81:43-49. [DOI] [PubMed] [Google Scholar]
- 133.Forst, S., J. Delgado, G. Ramakrishnan, and M. Inouye. 1988. Regulation of ompC and ompF expression in Escherichia coli in the absence of envZ. J. Bacteriol. 170:5080-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Forst, S., J. Gelgado, A. Rampersaud, and M. Inouye. 1990. In vivo phosphorylation of OmpR, the transcription activator of the ompF and ompC genes in Escherichia coli. J. Bacteriol. 172:3473-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fox, D. K., N. D. Meadow, and S. Roseman. 1986. Phosphate transfer between acetate kinase and enzyme I of the bacterial phosphotransferase system. J. Biol. Chem. 261:13498-13503. [PubMed] [Google Scholar]
- 136.Fox, D. K., and S. Roseman. 1986. Isolation and characterization of homogeous acetate kinase from Salmonella typhimurium and Escherichia coli. J. Biol. Chem. 261:13487-13497. [PubMed] [Google Scholar]
- 137.Fraenkel, D. G. 1996. Glycolysis, p. 189-198. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
- 138.Francez-Charlot, A., B. Laugel, A. Van Gemert, N. Dubarry, F. Wiorowski, M.-P. Castanie-Cornet, C. Gutierrez, and K. Cam. 2003. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol. Microbiol. 49:823-832. [DOI] [PubMed] [Google Scholar]
- 139.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, et al. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 140.Fraser, G. M., L. Claret, R. Furness, S. Gupta, and C. Hughes. 2002. Swarming- coupled expression of the Proteus mirabilis hpmBA haemolysin operon. Microbiology 148:2191-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Freestone, P., S. Grant, M. Trinei, T. Onoda, and V. Norris. 1998. Protein phosphorylation in Escherichia coli L. form NC-7. Microbiology 144:3289-3295. [DOI] [PubMed] [Google Scholar]
- 142.Freter, R. 1988. Mechanisms of bacterial colonization of the mucosal surfaces of the gut, p. 45-60, Virulence mechanisms of bacterial pathogens. American Society for Microbiology, Washington, D.C.
- 143.Freter, R. 1983. Mechanisms that control the microflora in the large intestine, p. 33-54. In D. J. Hentges (ed.), Human intestinal microflora in health and disease. Academic Press, Inc., New York, N.Y.
- 144.Fujino, T., J. Kondo, M. Ishikawa, K. Morikawa, and T. T. Yamamoto. 2001. Acetyl-CoA Synthetase 2, a mitochondrial matrix enzyme involved in the oxidation of acetate. J. Biol. Chem. 276:11420-11426. [DOI] [PubMed] [Google Scholar]
- 145.Galinier, A., J. Haiech, M. Kilhoffer, M. Jaquinod, J. Stulke, J. Deutscher, and I. Martin-Verstraete. 1997. The Bacillus subtilis crh gene encodes a HPr-like protein involved in carbon catabolite repression. Proc. Natl. Acad. Sci. USA 94:8439-8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Galinier, A., M. Kranaja, R. Engelmann, W. Hengstenber, M.-C. Kilhoffer, J. Deutscher, and J. Haiech. 1998. New protein kinase and protein phosphatase families mediate signal transduction in bacterial catabolite repression. Proc. Natl. Acad. Sci. USA 95:1823-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Galperin, M. Y., and N. V. Grishin. 2000. The synthetase domains of cobalamin biosynthesis amidotransferases cobB and cobQ belong to a new family of ATP-dependent amidoligases, related to dethiobiotin synthetase. Proteins 41:238-247. [DOI] [PubMed] [Google Scholar]
- 148.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 11:11-21. [DOI] [PubMed] [Google Scholar]
- 149.Garnak, M., and H. C. Reeves. 1979. Phosphorylation of isocitrate dehydrogenase of Escherichia coli. Science 203:1111-1112. [DOI] [PubMed] [Google Scholar]
- 150.Gennis, R. B., and L. P. Hager. 1976. Pyrvuate oxidase, p. 493-504. In A. N. Martonosi (ed.), The enzymes and biological membranes, vol. 2. Plenum, New York, N.Y. [Google Scholar]
- 151.Gerstmeir, R., A. Cramer, P. Dangel, S. Schaffer, and B. J. Eikmanns. 2004. RamB, a novel transcriptional regulator of genes involved in acetate metabolism of Corynebacterium glutamicum. J. Bacteriol. 186:2798-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gerstmeir, R., V. F. Wendisch, S. Schnicke, H. Ruan, M. Farwick, D. Reinscheid, and B. J. Eikmanns. 2003. Acetate metabolism and its regulation in Corynebacterium glutamicum. J. Biotechnol. 104:99-122. [DOI] [PubMed] [Google Scholar]
- 153.Gimenez, R., M. F. Nunez, J. Badia, J. Aguilar, and L. Baldoma. 2003. The gene yjcG, cotranscribed with the gene acs, encodes an acetate permease in Escherichia coli. J. Bacteriol. 185:6448-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Glasemacher, J., A. K. Bock, R. Schmid, and P. Schonheit. 1997. Purification and properties of acetyl-CoA synthetase (ADP-forming), an archaeal enzyme of acetate formation and ATP synthesis, from the hyperthermophile Pyrococcus furiosus. Eur. J. Biochem. 244:561-567. [DOI] [PubMed] [Google Scholar]
- 155.Gonzalez-Gil, G., R. Kahmann, and G. Muskhelishvili. 1998. Regulation of crp transcription by oscillation between distinct nucleoprotein complexes. EMBO J. 17:2877-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Goodier, R. I., and B. M. Ahmer. 2001. SirA orthologs affect both motility and virulence. J. Bacteriol. 183:2249-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gottschalk, G. 1985. Bacterial metabolism, 2nd ed. Springer-Verlag, New York, N.Y.
- 158.Gourse, R. L., W. Ross, and T. Gaal. 2000. UPs and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol. Microbiol. 37:687-695. [DOI] [PubMed] [Google Scholar]
- 159.Grabau, C., and J. E. J. Cronan. 1984. Molecular cloning of the gene (poxB) encoding the pyruvate oxidase of Escherichia coli, a lipid-activated enzyme. J. Bacteriol. 160:1088-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gray, C. T., J. W. T. Wimpenny, and W. R. Mossman. 1966. Regulation of metabolism in facultative bacteria. II. Effects of aerobiosis, anaerobiosis and nutrition on the formation of Kreb's cycle enzymes in Escherichia coli. Biochim. Biophys. Acta 117:33-41. [DOI] [PubMed] [Google Scholar]
- 161.Green, J., and M. Baldwin. 1997. HlyX, the FNR homologue of Actinobacillus pleuropneumoniae, is a [4Fe-4S]-containing oxygen-responsive transcription regulator that anaerobically activates FNR-dependent class I promoters via an enhanced AR1 contact. Mol. Microbiol. 24:593-605. [DOI] [PubMed] [Google Scholar]
- 162.Grozinger, C. M., and S. L. Schreiber. 2002. Deacetylase enzymes: biological functions and the use of small-molecule inhibitors. Chem. Biol. 9:3-16. [DOI] [PubMed] [Google Scholar]
- 163.Grundy, F. H., D. A. Waters, S. H. G. Allen, and T. M. Henkin. 1993. Identification of genes involved in utilization of acetate and acetoin in Bacillus subtilis. Mol. Microbiol. 10:259-271. [DOI] [PubMed] [Google Scholar]
- 164.Grundy, F. H., D. A. Waters, S. H. G. Allen, and T. M. Henkin. 1993. Regulation of the Bacillus subtilis acetate kinase gene by CcpA. J. Bacteriol. 175:7348-7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Grundy, F. J., A. J. Turinsky, and T. M. Henkin. 1994. Catabolite regulation of Bacillus subtilis acetate and acetoin utilization genes by CcpA. J. Bacteriol. 176:4527-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Guarente, L. 2000. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 14:1021-1026. [PubMed] [Google Scholar]
- 167.Guest, J. R., S. J. Angier, and G. C. Russell. 1989. Structure, expression, and protein engineering of the pyruvate dehydrogenase complex of Escherichia coli. Ann. N. Y. Acad. Sci. 573:76-99. [DOI] [PubMed] [Google Scholar]
- 168.Guest, J. R., and G. C. Russell. 1992. Complexes and complexities of the citric acid cycle in Escherichia coli. Curr. Top. Cell. Regul. 33:231-247. [DOI] [PubMed] [Google Scholar]
- 169.Gulick, A. M., V. J. Starai, A. R. Horswill, K. M. Homick, and J. C. Escalante- Semerena. 2003. The 1.75 A crystal structure of acetyl-CoA synthetase bound to adenosine-5′-propylphosphate and coenzyme A. Biochemistry 42:2866-2873. [DOI] [PubMed] [Google Scholar]
- 170.Gunsalus, R. P., and S. J. Park. 1994. Aerobic-anaerobic gene regulation in Escherichia coli: control by the ArcAB and Fnr regulons. Res. Microbiol. 145:437-450. [DOI] [PubMed] [Google Scholar]
- 171.Gupta, S., and D. P. Clark. 1989. Escherichia coli derivatives lacking both alcohol dehydrogenase and phosphotransacetylase grow anaerobically by lactate fermentation. J. Bacteriol. 171:3650-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hahm, D. H., J. Pan, and J. S. Rhee. 1994. Characterization and evaluation of a pta (phosphotransacetylase) negative mutant of Escherichia coli HB101 as production host of foreign lipase. Appl. Microbiol. Biotechnol. 42:100-107. [DOI] [PubMed] [Google Scholar]
- 173.Haldimann, A., S. Fisher, L. Daniels, C. Walsh, and B. Wanner. 1997. Transcriptional regulation of the Enterococcus faecium BM4147 vancomycin resistance gene cluster by the VanS-VanR two-component regulatory system in Escherichia coli K- 12. J. Bacteriol. 179:5903-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Han, K., H. C. Lim, and J. Hong. 1992. Acetic acid formation in Escherichia coli fermentation. Biotechnol. Bioeng. 39:663-671. [DOI] [PubMed] [Google Scholar]
- 175.Han, M.-J., S. S. Yoon, and S. Y. Lee. 2001. Proteome analysis of metabolically engineered Escherichia coli producing poly(3-hydroxybutyrate). J. Bacteriol. 183:301-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hansen, R. G., and U. Henning. 1966. Regulation of pyruvate dehydrogenase activity in Escherichia coli K12. Biochim. Biophys. Acta 122:355-358. [DOI] [PubMed] [Google Scholar]
- 177.Hansen, T., and P. Schonheit. 2000. Purification and properties of the first-identified, archaeal, ATP-dependent 6-phosphofructokinase, an extremely thermophilic non- allosteric enzyme, from the hyperthermophile Desulfurococcus amylolyticus. Arch. Microbiol. 173:103-109. [DOI] [PubMed] [Google Scholar]
- 178.Hanson, T. S., V. R. Srinivasan, and H. O. Halvorson. 1963. Biochemistry of sporulation. I. Metabolism of acetate by vegetative and sporulating cells. J. Bacteriol. 85:451-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Harms, C., U. Ludwig, and J. R. Andreesen. 1998. Sarcosine reductase of Tissierella creatinophila: purification and characterization of its components. Arch. Microbiol. 170:442-450. [DOI] [PubMed] [Google Scholar]
- 180.Hasty, P. 2001. The impact energy metabolism and genome maintenance have on longevity and senescence: lessons from yeast to mammals. Mech. Ageing Dev. 122:1651-1662. [DOI] [PubMed] [Google Scholar]
- 181.Head, C. G., A. Tardy, and L. J. Kenney. 1998. Relative binding affinities of OmpR and OmpR-phosphate at the ompF and ompC regulatory sites. J. Mol. Biol. 281:857-870. [DOI] [PubMed] [Google Scholar]
- 182.Heermann, R., K. Altendorf, and K. Jung. 2003. The N-terminal input domain of the sensor kinase KdpD of Escherichia coli stabilizes the interaction between the cognate response regulator KdpE and the corresponding DNA-binding site. J. Biol. Chem. 278:51277-51284. [DOI] [PubMed] [Google Scholar]
- 183.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the sigmaS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Henkin, T. M. 1996. The role of CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis. FEMS Microbiol. Lett. 135:9-15. [DOI] [PubMed] [Google Scholar]
- 185.Henkin, T. M., F. J. Grundy, W. L. Nicholson, and G. H. Chambliss. 1991. Catabolite repression of the α-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacI and galR repressors. Mol. Microbiol. 5:575-584. [DOI] [PubMed] [Google Scholar]
- 186.Hesslinger, C., S. A. Fairhurst, and G. Sawers. 1998. Novel keto acid formate-lyase and propionate kinase enzymes are components of an anaerobic pathway in Escherichia coli that degrades L-threonine to propionate. Mol. Microbiol. 27:477-492. [DOI] [PubMed] [Google Scholar]
- 187.Heyde, M., P. Laloi, and R. Portalier. 2000. Involvement of carbon source and acetyl phosphate in the external-pH-dependent expression of porin genes in Escherichia coli. J. Bacteriol. 182:198-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hickey, M. W., A. J. Hillier, and G. R. Jago. 1983. Metabolism of pyruvate and citrate in lactobacilli. Aust. J. Biol. Sci. 36:487-496. [DOI] [PubMed] [Google Scholar]
- 189.Hiesinger, M., C. Wagner, and H.-J. Schuller. 1997. The acetyl-CoA synthetase gene ACS2 of the yeast Saccharomyces cerevisiae is coregulated with structural genes of fatty acid biosynthesis by the transcriptional activators Ino2p and Ino4p. FEBS Lett. 415:16-20. [DOI] [PubMed] [Google Scholar]
- 190.Hiratsu, K., A. Nakata, H. Shinagawa, and K. Makino. 1995. Autophosphorylation and activation of transcriptional activator PhoB of Escherichia coli by acetyl phosphate in vitro. Gene 161:7-10. [DOI] [PubMed] [Google Scholar]
- 191.Hoch, J. A., and Silhavy, T. J. 1995. Two-component signal transduction. ASM Press, Washington, D.C.
- 192.Hoffman, T., N. Frankenberg, M. Marino, and D. Jahn. 1998. Ammonification in Bacillus subtilis utilizing dissimilatory nitrite reductase is dependent on resDE. J. Bacteriol. 180:186-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hollywood, N., and H. W. Doelle. 1976. Effect of specific growth rate and glucose concentration on growth and glucose metabolism of Escherichia coli K-12. Microbios 17:23-33. [PubMed] [Google Scholar]
- 194.Holman, T. R., Wu, Z., Wanner, B. L., and Walsh, C. T. 1994. Identification of the DNA-binding site for the phosphorylated VanR protein required for vancomycin resistance in Enterococcus faecium. Biochemistry 33:4625-4631. [DOI] [PubMed] [Google Scholar]
- 195.Holms, H. 1996. Flux analysis and control of the central metabolic pathways in Escherichia coli. FEMS Microbiol. Rev. 19:85-116. [DOI] [PubMed] [Google Scholar]
- 196.Holms, W. H. 1986. The central metabolic pathways of Escherichia coli: relationship between flux and control at a branch point, efficiency of conversion to biomass, and excretion of acetate. Curr. Top. Cell. Regul. 28:59-105. [DOI] [PubMed] [Google Scholar]
- 197.Hong, J.-S., and A. G. Hunt. 1980. The role of acetylphosphate in active transport. J. Supramol. Struct. 4:77. [Google Scholar]
- 198.Hong, J. S., A. G. Hunt, P. S. Masters, and M. A. Lieberman. 1979. Requirements of acetyl phosphate for the binding protein-dependent transport systems in Escherichia coli. Proc. Natl. Acad. Sci. USA 76:1213-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hormann, K., and J. R. Andereesen. 1989. Reductive cleavage of sarcosine and betaine by Eubacterium acidaminophilum via enzyme systems different from glycine reductase. Arch. Microbiol. 153:50-59. [Google Scholar]
- 200.Horswill, A. R., and J. C. Escalante-Semerena. 2002. Characterization of the propionyl-CoA synthetase (PrpE) enzyme of Salmonella enterica: residue Lys592 is required for propionyl-AMP synthesis. Biochemistry 41:2379-2387. [DOI] [PubMed] [Google Scholar]
- 201.Horswill, A. R., and J. C. Escalante-Semerena. 1999. The prpE gene of Salmonella typhimurium LT2 encodes propionyl-CoA synthetase. Microbiology 145:1381-1388. [DOI] [PubMed] [Google Scholar]
- 202.Horton, J. D., J. L. Goldstein, and M. S. Brown. 2002. SREBPs: transcriptional mediators of lipid homeostasis. Cold Spring Harbor Symp. Quant. Biol. 67:491-498. [DOI] [PubMed] [Google Scholar]
- 203.Hoyt, J., and H. Reeves. 1988. In vivo phosphorylation of isocitrate lyase from Escherichia coli. Biochem. Biophys. Res. Commun. 153:875-880. [DOI] [PubMed] [Google Scholar]
- 204.Huang, M., F. B. Oppermann-Sanio, and A. Steinbuchel. 1999. Biochemical and molecular characterization of the Bacillus subtilis acetoin catabolic pathway. J. Bacteriol. 181:3837-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Hubschmann, T., H. J. M. M. Jorissen, T. Borner, W. Gartner, and N. Tandeau de Marsac. 2001. Phosphorylation of proteins in the light-dependent signalling pathway of a filamentous cyanobacterium. Eur. J. Biochem. 268:3383-3389. [DOI] [PubMed] [Google Scholar]
- 206.Hueck, C. J., and W. Hillen. 1995. Catabolite repression in Bacillus subtilis: a global mechanism for the gram-positive bacteria? Mol. Microbiol. 15:395-401. [DOI] [PubMed] [Google Scholar]
- 207.Hueck, C. J., W. Hillen, and M. H. Saier, Jr. 1994. Analysis of a cis-active sequence mediating catabolite repression in gram-positive bacteria. Res. Microbiol. 145:503-518. [DOI] [PubMed] [Google Scholar]
- 208.Hunt, A. G. 1982. The energetics of osmotic shock-sensitive active transport in Escherichia coli: studies in whole cells and isolated membrane vesicles. PhD thesis. Brandeis, Waltham, Mass.
- 209.Hunt, A. G., and J.-S. Hong. 1983. Properties and characterization of binding protein dependent active transport of glutamine in isolated membrane vesicles of Escherichia coli. Biochemistry 22:844-850. [DOI] [PubMed] [Google Scholar]
- 210.Ikeda, Y., J. Yamamoto, M. Okamura, T. Fujino, S. Takahashi, K. Takeuchi, T. F. Osborne, T. T. Yamamoto, S. Ito, and J. Sakai. 2001. Transcriptional regulation of the murine acetyl-CoA synthetase 1 gene through multiple clustered binding sites for sterol regulatory element-binding proteins and a single neighboring site for Sp1. J. Biol. Chem. 276:34259-34269. [DOI] [PubMed] [Google Scholar]
- 211.Inouye, M., R. Dutta, and Y. Zhu. 2002. Regulation of porins in Escherichia coli by the osmosensing histidine kinase/phosphatase EnvZ, p. 25-46. In M. Inouye and R. Dutta (ed.), Histidine kinases in signal transduction. Academic Press, Ltd., London, United Kingdom.
- 212.Iuchi, S., and E. C. Lin. 1988. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc. Natl. Acad. Sci. USA 85:1888-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Jackowski, S. 1996. Biosynthesis of pantothenic acid and coenzyme A, p. 687-694. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American ASM Press, Washington, D.C.
- 214.Jackowski, S., and C. O. Rock. 1986. Consequences of reduced intracellular coenzyme A content in Escherichia coli. J. Bacteriol. 166:866-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Jackowski, S., and C. O. Rock. 1984. Metabolism of 4′-phosphopantetheine in Escherichia coli. J. Bacteriol. 158:115-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Jackowski, S., and C. O. Rock. 1981. Regulation of coenzyme A biosynthesis. J. Bacteriol. 148:926-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Janausch, I. G., I. Garcia-Moreno, D. Lehnen, Y. Zeuner, and G. Unden. 2004. Phosphorylation and DNA binding of the regulator DcuR of the fumarate-responsive two-component system DcuSR of Escherichia coli. Microbiology 150:877-883. [DOI] [PubMed] [Google Scholar]
- 218.Janiak-Spens, F., J. M. Sparling, M. Gurfinkel, and A. H. West. 1999. Differential stabilities of phosphorylated response regulator domains reflect functional roles of the yeast osmoregulatory SLN1 and SSK1 proteins. J. Bacteriol. 181:411-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Jensen, E. B., and S. Carlsen. 1990. Production of recombinant human growth hormone in Escherichia coli. Expression of different precursors and physiological effects of glucose, acetate and salts. Biotechnol. Bioeng. 36:1-11. [DOI] [PubMed] [Google Scholar]
- 220.Johannes, E., D. M. Barnhart, and J. L. Slonczewski. 2004. pH-dependent catabolic protein expression during anaerobic growth of Escherichia coli. J. Bacteriol. 186:192-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Jolly, C. A., H. Chao, A. B. Kier, J. T. Billheimer, and F. Schroeder. 2000. Sterol carrier protein-2 suppresses microsomal acyl-CoA hydrolysis. Mol. Cell. Biochem. 205:83-90. [DOI] [PubMed] [Google Scholar]
- 222.Jones, B. E., V. Dossonnet, E. Kuster, W. Hillen, J. Deutscher, and R. E. Klevit. 1997. Binding of the catabolite repressor protein CcpA to its DNA target is regulated by phosphorylation of its corepressor HPr. J. Biol. Chem. 272:26530-26535. [DOI] [PubMed] [Google Scholar]
- 223.Kaiser, M., and G. Sawers. 1994. Pyruvate formate-lyase is not essential for nitrate respiration by Escherichia coli. FEMS Microbiol. Lett. 117:163-168. [DOI] [PubMed] [Google Scholar]
- 224.Kakuda, H., K. Hosono, K. Shiroishi, and S. Ichihara. 1994. Identification and characterization of the ackA (acetate kinase A)-pta (phosphotransacetylase) operon and complementation analysis of acetate utilization by an ackA-pta deletion mutant of Escherichia coli. J. Biochem. 116:916-922. [DOI] [PubMed] [Google Scholar]
- 225.Kakuda, H., K. Shiroishi, K. Hosono, and S. Ichihara. 1994. Construction of Pta-Ack pathway deletion mutants of Escherichia coli and characteristic growth profiles of the mutants in a rich medium. Biosci. Biotechnol. Biochem. 58:2232-2235. [DOI] [PubMed] [Google Scholar]
- 226.Kao, K. C., Y.-L. Yang, R. Boscolo, C. Sabatti, V. Roychowdhury, and J. C. Liao. 2004. Transcriptome-based determination of multiple transcription regulator activities in Escherichia coli by using network component analysis. Proc. Natl. Acad. Sci. USA 101:641-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Karan, D., J. R. David, and P. Capy. 2001. Molecular evolution of the AMP-forming Acetyl-CoA synthetase. Gene 265:95-101. [DOI] [PubMed] [Google Scholar]
- 228.Kasahara, M., and M. Ohmori. 1999. Activation of a cyanobacterial adenylate cyclase, CyaC, by autophosphorylation and a subsequent phosphotransfer reaction. J. Biol. Chem. 274:15167-15172. [DOI] [PubMed] [Google Scholar]
- 229.Kaya, S., T. Yokoyama, Y. Hayashi, K. Taniguchi, and T. Tsuda. 1998. ATP and acetyl phosphate induces molecular events near the ATP binding site and the membrane domain of Na+,K+-ATPase. The tetrameric nature of the enzyme. J. Biol. Chem. 273:24334-24338. [DOI] [PubMed] [Google Scholar]
- 230.Ke, J., R. H. Behal, S. L. Back, B. J. Nikolau, E. S. Wurtele, and D. J. Oliver. 2000. The role of pyruvate dehydrogenase and acetyl-coenzyme a synthetase in fatty acid synthesis in developing arabidopsis seeds. Plant Physiol. 123:497-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Kennedy, E. P. 2001. Hitler's gift and the era of biosynthesis. J. Biol. Chem. 276:42619-42631. [DOI] [PubMed] [Google Scholar]
- 232.Kessler, D., and J. Knappe. 1996. Anaerobic dissimilation of pyruvate, p. 199-204. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 233.Kihara, M., and R. M. Macnab. 1981. Cytoplasmic pH mediates pH taxis and weak- acid repellent taxis of bacteria. J. Bacteriol. 145:1209-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Kim, C. C., and S. Falkow. 2004. Delineation of upstream signaling events in the salmonella pathogenicity island 2 transcriptional activation pathway. J. Bacteriol. 186:4694-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Kim, D.-J., B. Boylan, N. George, and S. Forst. 2003. Inactivation of ompR promotes precocious swarming and flhDC expression in Xenorhabdus nematophila. J. Bacteriol. 185:5290-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Kim, J. H., Z. T. Guvener, J. Y. Cho, K. Chung, and G. H. Chambliss. 1995. Specificity of DNA binding activity of the Bacillus subtilis catabolite control protein CcpA. J. Bacteriol. 177:5129-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Kim, S. B., B. S. Shin, S. K. Choi, C. K. Kim, and S. H. Park. 2001. Involvement of acetyl phosphate in the in vivo activation of the response regulator ComA in Bacillus subtilis. FEMS Microbiol. Lett. 195:179-183. [DOI] [PubMed] [Google Scholar]
- 238.Kim, S. K., M. R. Wilmes-Riesenberg, and B. L. Wanner. 1996. Involvement of the sensor kinase EnvZ in the in vivo activation of the response-regulator PhoB by acetyl phosphate. Mol. Microbiol. 22:135-147. [DOI] [PubMed] [Google Scholar]
- 239.Kimata, K., H. Takahashi, T. Inada, P. Postma, and H. Aiba. 1997. cAMP receptor protein-cAMP plays a crucial role in glucose-lactose diauxie by activating the major glucose transporter gene in Escherichia coli. Proc. Natl. Acad. Sci. USA 94:12914-12919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Kirkpatrick, C., L. M. Maurer, N. E. Oyelakin, Y. N. Yoncheva, R. Maurer, and J. L. Slonczewski. 2001. Acetate and formate stress: opposite responses in the proteome of Escherichia coli. J. Bacteriol. 183:6466-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Kleman, G. L., and W. R. Strohl. 1994. Acetate metabolism by Escherichia coli in high-cell-density fermentation. Appl. Environ. Microbiol. 60:3952-3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Knappe, J., and G. Sawers. 1990. A radical-chemical route to acetyl-CoA: the anerobically induced pyruvate formate-lyase system of Escherichia coli. FEMS Microbiol. Rev. 75:383-398. [DOI] [PubMed] [Google Scholar]
- 243.Knorr, R., M. A. Ehrmann, and R. F. Vogel. 2001. Cloning of the phosphotransacetylase gene from Lactobacillus sanfranciscensis and characterization of its gene product. J. Basic Microbiol. 41:339-349. [DOI] [PubMed] [Google Scholar]
- 244.Knorr, R., M. A. Ehrmann, and R. F. Vogel. 2001. Cloning, expression, and characterization of acetate kinase from Lactobacillus sanfranciscensis. Microbiol. Res. 156:267-277. [DOI] [PubMed] [Google Scholar]
- 245.Knudsen, J., M. V. Jensen, J. K. Hansen, N. J. Faergeman, T. B. Neergaard, and B. Gaigg. 1999. Role of acylCoA binding protein in acylCoA transport, metabolism and cell signaling. Mol. Cell. Biochem. 192:95-103. [DOI] [PubMed] [Google Scholar]
- 246.Kornberg, H. L. 1966. The role and control of the glyoxylate cycle in Escherichia coli. Biochem. J. 99:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Kouzarides, T. 2000. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 19:1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Kravanja, M., R. Engelmann, V. Dossonnet, M. Bluggel, H. E. Meyer, R. Frank, A. Galinier, J. Deutscher, N. Schnell, and W. Hengstenberg. 1999. The hprK gene of Enterococcus faecalis encodes a novel bifunctional enzyme: the HPr kinase/phosphatase. Mol. Microbiol. 31:59-66. [DOI] [PubMed] [Google Scholar]
- 249.Kumari, S., C. M. Beatty, D. F. Browning, S. J. Busby, E. J. Simel, G. Hovel-Miner, and A. J. Wolfe. 2000. Regulation of acetyl coenzyme A synthetase in Escherichia coli. J. Bacteriol. 182:4173-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Kumari, S., E. Simel, and A. J. Wolfe. 2000. Sigma70 is the principal sigma factor responsible for the transcription of acs, which encodes acetyl-CoA synthetase in Escherichia coli. J. Bacteriol. 182:551-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Kumari, S., R. Tishel, M. Eisenbach, and A. J. Wolfe. 1995. Cloning, characterization, and functional expression of acs, the gene which encodes acetyl coenzyme A synthetase in Escherichia coli. J. Bacteriol. 177:2878-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, et al. 1998. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 253.Kuster, S., E. J. Luesink, W. M. de Vos, and W. Hillen. 1996. Immunological crossreactivity to catabolite control protein CcpA from Bacillus megaterium is found in many gram-positive bacteria. FEMS Microbiol. Lett. 139:109-115. [DOI] [PubMed] [Google Scholar]
- 254.Kwan, H. S., H. W. Chui, and K. K. Wong. 1988. ack::Mu d1-8 (Apr lac) operon fusions of Salmonella typhimurium LT-2. Mol. Gen. Genet. 211:183-185. [DOI] [PubMed] [Google Scholar]
- 255.Kwon, Y. M., and S. C. Ricke. 1998. Induction of acid resistance of Salmonella typhimurium by exposure to short-chain fatty acids. Appl. Environ. Microbiol. 64:3458-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Lambert, L., K. Abshire, D. Blankenhorn, and J. Slonczewski. 1997. Proteins induced in Escherichia coli by benzoic acid. J. Bacteriol. 179:7595-7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.LaPorte, D., P. Thorness, and D. Koshland, Jr. 1985. Compensatory phosphorylation of isocitrate dehydrogenase, a mechanism for adaption to the intracellular environment. J. Biol. Chem. 260:10563-10568. [PubMed] [Google Scholar]
- 258.LaPorte, D. C., and D. E. Koshland, Jr. 1983. Phosphorylation of isocitrate dehydrogenase as a demonstration of enhanced sensitivity in covalent regulation. Nature 305:286-290. [DOI] [PubMed] [Google Scholar]
- 259.Latimer, M. T., and J. G. Ferry. 1993. Cloning, sequence analysis, and hyperexpression of the genes encoding phosphotransacetylase and acetate kinase from Methanosarcina thermophila. J. Bacteriol. 175:6822-6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Lawhon, S. D., R. Maurer, M. Suyemoto, and C. Altier. 2002. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol. Microbiol. 46:451-464. [DOI] [PubMed] [Google Scholar]
- 261.Lee, A. K., C. S. Detweiler, and S. Falkow. 2000. Ompr regulates the two-component system SsrA-SsrB in Salmonella pathogenicity island 2. J. Bacteriol. 182:771-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Lee, J.-H., D.-E. Lee, B.-U. Lee, and H.-S. Kim. 2003. Global analyses of transcriptomes and proteomes of a parent strain and an l-threonine-overproducing mutant strain. J. Bacteriol. 185:5442-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Lee, S. Y. 1996. High cell density culture of Escherichia coli. Trends Biotechnol. 14:98-105. [DOI] [PubMed] [Google Scholar]
- 264.Lee, T.-Y., K. Makino, H. Shinagawa, and A. Nakata. 1990. Overproduction of acetate kinase activates the phosphate regulon in the absence of the phoR and phoM functions in Escherichia coli. J. Bacteriol. 172:2245-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Lehninger, A. L. 1975. Biochemistry, 2nd ed. Worth Publishers, Inc., New York, N.Y.
- 266.Lesley, J. A., and C. D. Waldburger. 2003. Repression of Escherichia coli PhoP-PhoQ signaling by acetate reveals a regulatory role for acetyl coenzyme A. J. Bacteriol. 185:2563-2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Leuchtenberger, W. 1996. Amino acids—technical production and use, p. 465-502. In H. J. Rehm, G. Reed, A. Puhler, P. Stadler, and M. Roehr (ed.), Biotechnology, vol. 6. VCH Verlagsgesellschaft, Weinheim, Germany. [Google Scholar]
- 268.Li, J., S. Kustu, and V. Stewart. 1994. In vitro interaction of nitrate-responsive regulatory protein NarL with DNA target sequences in the fdnG, narG, narK and frdA operon control regions of Escherichia coli K-12. J. Mol. Biol. 241:150-165. [DOI] [PubMed] [Google Scholar]
- 269.Liao, J. D., S.-Y. Hou, and Y.-P. Chao. 1996. Pathway analysis, engineering, and physiological considerations for redirecting central metabolism. Biotechnol. Bioeng. 52:129-140. [DOI] [PubMed] [Google Scholar]
- 270.Lin, J., I. S. Lee, J. Frey, J. L. Slonczewski, and J. W. Foster. 1995. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J. Bacteriol. 177:4097-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Lipmann, F. 1941. Metabolic generation and utilization of phosphate bond energy. Adv. Enzymol. 1:99-162.
- 272.Liu, J. H., M. J. Lai, S. Ang, J. C. Shu, P. C. Soo, Y. T. Horng, W. C. Yi, H. C. Lai, K. T. Luh, S. W. Ho, and S. Swift. 2000. Role of flhDC in the expression of the nuclease gene nucA, cell division and flagellar synthesis in Serratia marcescens. J. Biomed. Sci. 7:475-483. [DOI] [PubMed] [Google Scholar]
- 273.Liu, X., and T. Ferenci. 2001. An analysis of multifactorial influences on the transcriptional control of ompF and ompC porin expression under nutrient limitation. Microbiology 147:2981-2989. [DOI] [PubMed] [Google Scholar]
- 274.Liu, X., and T. Ferenci. 1998. Regulation of porin-mediated outer membrane permeability by nutrient limitation in Escherichia coli. J. Bacteriol. 180:3917-3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Loh, J., M. Garcia, and G. Stacey. 1997. NodV and NodW, a second flavonoid recognition system regulating nod gene expression in Bradyrhizobium japonicum. J. Bacteriol. 179:3013-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Loikkanen, I., S. Haghighi, S. Vainio, and A. Pajunen. 2002. Expression of cytosolic acetyl-CoA synthetase gene is developmentally regulated. Mech. Dev. 115:139-141. [DOI] [PubMed] [Google Scholar]
- 277.Luby-Phelps, K. 2000. Cytoarchitecture and physical properties of cytoplasm: volume, viscosity, diffusion, intracellular surface area. Int. Rev. Cytol. 192:189-221. [DOI] [PubMed] [Google Scholar]
- 278.Luby-Phelps, K. 1994. Physical properties of cytoplasm. Curr. Opin. Cell Biol. 6:3-9. [DOI] [PubMed] [Google Scholar]
- 279.Lukat, G. S., W. R. McCleary, A. M. Stock, and J. B. Stock. 1992. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc. Natl. Acad. Sci. USA 89:718-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Luli, G. W., and W. R. Strohl. 1990. Comparison of growth, acetate production, and acetate inhibition of Escherichia coli strains in batch and fed-batch fermentations. Appl. Environ. Microbiol. 56:1004-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Luong, A., V. C. Hannah, M. S. Brown, and J. L. Goldstein. 2000. Molecular characterization of human acetyl-CoA synthetase, an enzyme regulated by sterol regulatory element-binding proteins. J. Biol. Chem. 275:26458-26466. [DOI] [PubMed] [Google Scholar]
- 282.Lynch, A. S., and Lin, E. C. C. 1996. Responses to molecular oxygen, p. 1526-1538. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 283.Macfarlane, S., A. J. McBain, and G. T. Macfarlane. 1997. Consequences of biofilm and sessile growth in the large intestine. Adv. Dent. Res. 11:59-68. [DOI] [PubMed] [Google Scholar]
- 284.Mackie, R., A. Sghir, and H. R. Gaskins. 1999. Developmental microbial ecology of the neonatal gastrointestinal tract. Am. J. Clin. Nutr. 69:1035S-1045S. [DOI] [PubMed] [Google Scholar]
- 285.Mai, X., and M. Adams. 1996. Purification and characterization of two reversible and ADP-dependent acetyl coenzyme A synthetases from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 178:5897-5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Majewski, R. A., and M. M. Domach. 1990. Simple constrained optimization view of acetate overflow in E. coli. Biotechnol. Bioeng. 35:732-738. [DOI] [PubMed] [Google Scholar]
- 287.Makino, K., M. Amemura, S.-K. Kim, H. Shinagawa, and A. Hakata. 1992. Signal transduction of the phosphate regulon in Escherichia coli mediated by phosphorylation. p. 191-200. In S. Papa, A. Azzi, and J. M. Tager (ed.), Adenine nucleotides in cellular energy transfer and signal transduction. Birkhaeuser Verlag, Basel, Switzerland.
- 288.Makino, K., H. Shinagawa, M. Amemura, S. Kimura, A. Nakata, and A. Ishihama. 1988. Regulation of the phosphate regulon of Escherichia coli: activation of pstS transcription by PhoB protein in vitro. J. Mol. Biol. 203:85-95. [DOI] [PubMed] [Google Scholar]
- 289.Marino, M., H. C. Ramos, T. Hoffmann, P. Glaser, and D. Jahn. 2001. Modulation of anaerobic energy metabolism of Bacillus subtilis by arfM (ywiD). J. Bacteriol. 183:6815-6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Marshall, F. A., S. L. Messenger, N. R. Wyborn, J. R. Guest, H. Wing, S. J. Busby, and J. Green. 2001. A novel promoter architecture for microaerobic activation by the anaerobic transcription factor FNR. Mol. Microbiol. 39:747-753. [DOI] [PubMed] [Google Scholar]
- 291.Matsubara, M., and T. Mizuno. 1999. EnvZ-independent phosphotransfer signaling pathway of the OmpR-mediated osmoregulatory expression of OmpC and OmpF in Escherichia coli. Biosci. Biotechnol. Biochem. 63:408-414. [DOI] [PubMed] [Google Scholar]
- 292.Matsubara, M., and T. Mizuno. 2000. The SixA phospho-histidine phosphatase modulates the ArcB phosphorelay signal transduction in Escherichia coli. FEBS Lett. 470:118-124. [DOI] [PubMed] [Google Scholar]
- 293.Matsushika, A., and T. Mizuno. 1998. A dual-signaling mechanism mediated by the ArcB hybrid sensor kinase containing the histidine-containing phosphotransfer domain in Escherichia coli. J. Bacteriol. 180:3973-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Matsuyama, A., H. Yamamoto-Otake, J. Hewitt, R. T. A. MacGillivray, and E. Nakano. 1994. Nucleotide sequence of the phosphotransacetylase gene of Escherichia coli strain K12. Biochim. Biophys. Acta 1219:559-562. [DOI] [PubMed] [Google Scholar]
- 295.Mayover, T. L., C. J. Halkides, and R. C. Stewart. 1999. Kinetic characterization of CheY phosphorylation reactions: comparison of P-CheA and small-molecule phosphodonors. Biochemistry 38:2259-2271. [DOI] [PubMed] [Google Scholar]
- 296.McCleary, W. R. 1996. The activation of PhoB by acetylphosphate. Mol. Microbiol. 20:1155-1163. [DOI] [PubMed] [Google Scholar]
- 297.McCleary, W. R., and J. B. Stock. 1994. Acetyl phosphate and the activation of two- component response regulators. J. Biol. Chem. 269:31567-31572. [PubMed] [Google Scholar]
- 298.McCleary, W. R., J. B. Stock, and A. J. Ninfa. 1993. Is acetyl phosphate a global signal in Escherichia coli? J. Bacteriol. 175:2793-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.McLeod, S. M., and R. C. Johnson. 2001. Control of transcription by nucleoid proteins. Curr. Opin. Microbiol. 4:152-159. [DOI] [PubMed] [Google Scholar]
- 300.McNeil, N. I. 1984. The contribution of the large intestine to energy supplies in man. Am. J. Clin. Nutr. 39:338-342. [DOI] [PubMed] [Google Scholar]
- 301.Meile, L., L. M. Rohr, T. A. Geissmann, M. Herensperger, and M. Teuber. 2001. Characterization of the d-xylulose 5-phosphate/d-fructose 6-phosphate phosphoketolase gene (xfp) from Bifidobacterium lactis. J. Bacteriol. 183:2929-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Merkler, I., and J. Retey. 1981. Stereochemical investigation of the phosphoketolase reaction. The formation of chiral [2H1,3H]acetyl phosphate. Eur. J. Biochem. 120:593-597. [DOI] [PubMed] [Google Scholar]
- 303.Mevissen-Verhage, E. A. E., V. H. Marcelis, M. N. De Vos, W. C. M. Harmsen-Vann Amerongen, and J. Verhoef. 1987. Bfidobacterium, Bacteroides and Clostridium spp. in fecal samples from breast-fed and bottle-fed infants with and without iron supplement. J. Clin. Microbiol. 25:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Meyer, M., P. Dimroth, and M. Bott. 1997. In vitro binding of the response regulator CitB and of its carboxy-terminal domain to A + T-rich DNA target sequences in the control region of the divergent citC and citS operons of Klebsiella pneumoniae. J. Mol. Biol. 269:719-731. [DOI] [PubMed] [Google Scholar]
- 305.Meyer, M., K. Granderath, and J. R. Andreesen. 1995. Purification and characterization of protein PB of betaine reductase and its relationship to the corresponding proteins glycine reductase and sarcosine reductase from Eubacterium acidaminophilum. Eur. J. Biochem. 234:184-191. [DOI] [PubMed] [Google Scholar]
- 306.Mitsuoka, T. 1996. Intestinal flora and human health. Asia Pacific J. Clin. Nutr. 5:2-9. [PubMed] [Google Scholar]
- 307.Miwa, Y., M. Saikawa, and Y. Fujita. 1994. Possible function and some properties of the CcpA protein of Bacillus subtilis. Microbiology 140:2567-2575. [DOI] [PubMed] [Google Scholar]
- 308.Miyake, M., K. Kataoka, M. Shirai, and Y. Asada. 1997. Control of poly-beta- hydroxybutyrate synthase mediated by acetyl phosphate in cyanobacteria. J. Bacteriol. 179:5009-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Miyake, M., C. Miyamoto, J. Schnackenberg, R. Kurane, and Y. Asada. 2000. Phosphotransacetylase as a key factor in biological production of polyhydroxybutyrate. Appl. Biochem. Biotechnol. 84-86:1039-1044. [DOI] [PubMed] [Google Scholar]
- 310.Miyake, M., K. Takase, M. Narato, E. Khatipov, J. Schnackenberg, M. Shirai, R. Kurane, and Y. Asada. 2000. Polyhydroxybutyrate production from carbon dioxide by cyanobacteria. Appl. Biochem. Biotechnol. 84-86:991-1002. [DOI] [PubMed] [Google Scholar]
- 311.Moazed, D. 2001. Common themes in mechanisms of gene silencing. Mol. Cell 8:489-498. [DOI] [PubMed] [Google Scholar]
- 312.Moir-Blais, T. R., F. J. Grundy, and T. M. Henkin. 2001. Transcriptional activation of the Bacillus subtilis ackA promoter requires sequences upstream of the CcpA binding site. J. Bacteriol. 183:2389-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Mortensen, P. B., and M. R. Clausen. 1996. Short-chain fatty acids in the human colon: relation to gastrointestinal health and disease. Scand. J. Gastroenterol. Suppl. 216:132-148. [DOI] [PubMed] [Google Scholar]
- 314.Muffler, A., S. Bettermann, M. Haushalter, A. Horlein, U. Neveling, M. Schramm, and O. Sorgenfrei. 2002. Genome-wide transcription profiling of Corynebacterium glutamicum after heat shock and during growth on acetate and glucose. J. Biotechnol. 98:255-268. [DOI] [PubMed] [Google Scholar]
- 315.Murphy, M. G., L. O'Connor, D. Walsh, and S. Condon. 1985. Oxygen dependent lactate utilization by Lactobacillus plantarum. Arch. Microbiol. 141:75-79. [DOI] [PubMed] [Google Scholar]
- 316.Musfeldt, M., M. Selig, and P. Schonheit. 1999. Acetyl coenzyme A synthetase (ADP forming) from the hyperthermophilic archaeon Pyrococcus furiosus: identification, cloning, separate expression of the encoding genes, acdAI and acdBI, in Escherichia coli, and in vitro reconstitution of the active heterotetrameric enzyme from its recombinant subunits. J. Bacteriol. 181:5885-5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Nakano, M. M., Y. P. Dailly, P. Zuber, and D. P. Clark. 1997. Characterization of anaerobic fermentative growth in Bacillus subtilis: identification of fermentation end products and genes required for the growth. J. Bacteriol. 179:6749-6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Nakayama, S.-I., and H. Watanabe. 1998. Identification of cpxR as a positive regulator essential for expression of the Shigella sonnei virF gene. J. Bacteriol. 180:3522-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Negre, D., C. Oudot, J.-F. Prost, K. Murakami, A. Ishihama, A. J. Cozzone, and J.-C. Cortay. 1998. FruR-mediated transcriptional activation at the ppsA promoter of Escherichia coli. J. Mol. Biol. 276:355-365. [DOI] [PubMed] [Google Scholar]
- 320.Neidhardt, F. C., J. L. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell: a molecular approach. Sinauer Associates, Inc., Sunderland, Mass.
- 321.Ninfa, A. J. 1996. Regulation of gene transcription by extracellular stimuli. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella cellular and molecular biology. ASM Press, Washington, D.C.
- 322.Ninfa, A. J., P. Jiang, M. R. Atkinson, and J. A. Peliska. 2000. Integration of antagonistic signals in the regulation of nitrogen assimilation in Escherichia coli. Curr. Top. Cell Regul. 36:31-75. [DOI] [PubMed] [Google Scholar]
- 323.Noronha, S. B., H. J. Yeh, T. F. Spande, and J. Shiloach. 2000. Investigation of the TCA cycle and the glyoxylate shunt in Escherichia coli BL21 and JM109 using (13)C- NMR/MS. Biotechnol. Bioeng. 68:316-327. [PubMed] [Google Scholar]
- 324.Nyström, T. 1994. The glucose-starvation stimulon of Escherichia coli: induced and repressed synthesis of enzymes of central metabolic pathways and role of acetyl phosphate in gene expression and starvation survival. Mol. Microbiol. 12:833-843. [DOI] [PubMed] [Google Scholar]
- 325.Nyström, T., and F. C. Neidhardt. 1993. Isolation and properties of a mutant of Escherichia coli with an insertional inactivation of the uspA gene, which encodes a universal stress protein. J. Bacteriol. 175:3949-3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Ogino, T., Y. Arata, and S. Fujiwara. 1980. Proton correlation nuclear magnetic resonance study of metabolic regulations and pyruvate transport in anaerobic Escherichia coli cells. Biochemistry 19:3684-3691. [DOI] [PubMed] [Google Scholar]
- 327.Ogino, T., M. Matsubara, N. Kato, Y. Nakamura, and T. Mizuno. 1998. An Escherichia coli protein that exhibits phosphohistidine phosphatase activity towards the HPt domain of the ArcB sensor involved in the multistep His-Asp phosphorelay. Mol. Microbiol. 27:573-585. [DOI] [PubMed] [Google Scholar]
- 328.Oh, M. K., and J. C. Liao. 2000. Gene expression profiling by DNA microarrays and metabolic fluxes in Escherichia coli. Biotechnol. Prog. 16:278-286. [DOI] [PubMed] [Google Scholar]
- 329.Oh, M.-K., L. Rohlin, K. C. Kao, and J. C. Liao. 2002. Global expression profiling of acetate-grown Escherichia coli. J. Biol. Chem. 277:13175-13183. [DOI] [PubMed] [Google Scholar]
- 330.Oshima, T., H. Aiba, Y. Masuda, S. Kanaya, M. Sugiura, B. L. Wanner, H. Mori, and T. Mizuno. 2002. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol. Microbiol. 46:281-291. [DOI] [PubMed] [Google Scholar]
- 331.Palacios, S., V. J. Starai, and J. C. Escalante-Semerena. 2003. Propionyl coenzyme a is a common intermediate in the 1,2-propanediol and propionate catabolic pathways needed for expression of the prpBCDE operon during growth of Salmonella enterica on 1,2-Propanediol. J. Bacteriol. 185:2802-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Pan, J. G., J. S. Rhee, and J. M. Lebeault. 1987. Physiological constraints in increasing biomass concentration of Escherichia coli in fed-batch culture. Biotechnol. Lett. 9:89-94. [Google Scholar]
- 333.Pantazaki, A. A., M. G. Tambaka, V. Langlois, P. Guerin, and D. A. Kyriakidis. 2004. Polyhydroxyalkanoate (PHA) biosynthesis in Thermus thermophilus: purification and biochemical properties of PHA synthase. Mol. Cell. Biochem. 254:173-183. [DOI] [PubMed] [Google Scholar]
- 334.Pardee, A. B., and L. S. Prestidge. 1955. Induced formation of serine and threonine deaminases by Escherichia coli. J. Bacteriol. 70:667-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Park, S.-J., G. Chao, and R. P. Gunsalus. 1997. Aerobic regulation of the sucABCD genes of Escherichia coli, which encode alpha-ketoglutarate dehydrogenase and succinyl coenzyme A synthetase: roles of ArcA, Fnr, and the upstream sdhCBAB promoter. J. Bacteriol. 179:4138-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Park, S. J., P. A. Cotter, and R. P. Gunsalus. 1995. Regulation of malate dehydrogenase (mdh) gene expression in Escherichia coli in response to oxygen, carbon, and heme availability. J. Bacteriol. 177:6652-6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Park, S. J., and R. P. Gunsalus. 1995. Oxygen, iron, carbon, and superoxide control of the fumarase fumA and fumC genes of Escherichia coli: role of the arcA, fnr, and soxR gene products. J. Bacteriol. 177:6255-6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Park, S. J., J. McCabe, J. Turna, and R. P. Gunsalus. 1994. Regulation of the citrate synthase (gltA) gene of Escherichia coli in response to anaerobiosis and carbon supply: role of the arcA gene product. J. Bacteriol. 176:5086-5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Park, S. J., C. P. Tseng, and R. P. Gunsalus. 1995. Regulation of succinate dehydrogenase (sdhCDAB) operon expression in Escherichia coli in response to carbon supply and anaerobiosis: role of ArcA and Fnr. Mol. Microbiol. 15:473-482. [DOI] [PubMed] [Google Scholar]
- 340.Patnaik, R., W. D. Roof, R. F. Young, and J. C. Liao. 1992. Stimulation of glucose catabolism in Escherichia coli by a potential futile cycle. J. Bacteriol. 174:7527-7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Peekhaus, N., and T. Conway. 1998. What's for dinner? Entner-Doudoroff metabolism in Escherichia coli. J. Bacteriol. 180:3495-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Peng, L., and K. Shimizu. 2003. Global metabolic regulation analysis for Escherichia coli K12 based on protein expression by 2-dimensional electrophoresis and enzyme activity measurement. Appl. Microbiol. Biotechnol. 61:163-178. [DOI] [PubMed] [Google Scholar]
- 343.Pericone, C. D., S. Park, J. A. Imlay, and J. N. Weiser. 2003. Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the Fenton reaction. J. Bacteriol. 185:6815-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Perrot, F., M. Hebraud, R. Charlionet, G. Junter, and T. Jouenne. 2000. Protein patterns of gel-entrapped Escherichia coli cells differ from those of free-floating organisms. Electrophoresis 21:645-653. [DOI] [PubMed] [Google Scholar]
- 345.Pettijohn, D. 1996. The nucleoid, p. 158-166. In F. C. Neidhardt, R. Curtiss, III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 346.Phadtare, S., and M. Inouye. 2001. Role of CspC and CspE in regulation of expression of RpoS and UspA, the stress response proteins in Escherichia coli. J. Bacteriol. 183:1205-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Phue, J.-N., and J. Shiloach. 2004. Transcription levels of key metabolic genes are the cause for different glucose utilization pathways in E. coli B (BL21) and E. coli K (JM109). J. Biotechnol. 109:21-30. [DOI] [PubMed] [Google Scholar]
- 348.Pittman, M. S., M. Goodwin, and D. J. Kelly. 2001. Chemotaxis in the human gastric pathogen Helicobacter pylori: different roles for CheW and the three CheV paralogues, and evidence for CheV2 phosphorylation. Microbiology 147:2493-2504. [DOI] [PubMed] [Google Scholar]
- 349.Plumbridge, J. 2002. Regulation of gene expression in the PTS in Escherichia coli: the role and interactions of Mlc. Curr. Opin. Microbiol. 5:187-193. [DOI] [PubMed] [Google Scholar]
- 350.Posthuma, C. C., R. Bader, R. Engelmann, P. W. Postma, W. Hengstenberg, and P. H. Pouwels. 2002. Expression of the xylulose 5-phosphate phosphoketolase gene, xpkA, from Lactobacillus pentosus MD363 is induced by sugars that are fermented via the phosphoketolase pathway and is repressed by glucose mediated by CcpA and the mannose phosphoenolpyruvate phosphotransferase system. Appl. Environ. Microbiol. 68:831-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1996. Phosphoenolpyruvate:carbohydrate phosphotransferase systems, p. 1149-1174. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 352.Poulsen, L., T. Licht, C. Rang, K. Krogfelt, and S. Molin. 1995. Physiological state of Escherichia coli BJ4 growing in the large intestines of streptomycin-treated mice. J. Bacteriol. 177:5840-5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Presecan-Siedel, E., A. Galinier, R. Longin, J. Deutscher, A. Danchin, P. Glaser, and I. Martin-Verstraete. 1999. Catabolite regulation of the pta gene as part of carbon flow pathways in Bacillus subtilis. J. Bacteriol. 181:6889-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Prohinar, P., S. A. Forst, D. Reed, I. Mandic-Mulec, and J. Weiss. 2002. OmpR- dependent and OmpR-independent responses of Escherichia coli to sublethal attack by the neutrophil bactericidal/permeability increasing protein. Mol. Microbiol. 43:1493-1504. [DOI] [PubMed] [Google Scholar]
- 355.Prüß, B. M. 1998. Acetyl phosphate and the phosphorylation of OmpR are involved in the regulation of the cell division rate in Escherichia coli. Arch. Microbiol. 170:141-146. [DOI] [PubMed] [Google Scholar]
- 356.Prüß, B. M., J. W. Campbell, T. K. Van Dyk, C. Zhu, Y. Kogan, and P. Matsumura. 2003. FlhD/FlhC Is a regulator of anaerobic respiration and the Entner-Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer. J. Bacteriol. 185:534-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Prüß, B. M., X. Liu, W. Hendrickson, and P. Matsumura. 2001. FlhD/FlhC-regulated promoters analyzed by gene array and lacZ gene fusions. FEMS Microbiol. Lett. 197:91-97. [DOI] [PubMed] [Google Scholar]
- 358.Prüß, B. M., and P. Matsumura. 1996. A regulator of the flagellar regulon of Escherichia coli, flhD, also affect cell division. J. Bacteriol. 178:668-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Prüß, B. M., J. M. Nelms, C. Park, and A. J. Wolfe. 1994. Mutations in NADH:ubiquinone oxidoreductase of Escherichia coli affect growth on mixed amino acids. J. Bacteriol. 176:2143-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Prüß, B. M., and A. J. Wolfe. 1994. Regulation of acetyl phosphate synthesis and degradation, and the control of flagellar expression in Escherichia coli. Mol. Microbiol. 12:973-984. [DOI] [PubMed] [Google Scholar]
- 361.Puchowicz, M. A., I. R. Bederman, B. Comte, D. Yang, F. David, E. Stone, K. Jabbour, D. H. Wasserman, and H. Brunengraber. 1999. Zonation of acetate labeling across the liver: implications for studies of lipogenesis by MIDA. Am. J. Physiol. Endocrinol. Metab. 277:E1022-E1027. [DOI] [PubMed] [Google Scholar]
- 362.Quail, M. A., D. J. Haydon, and J. R. Guest. 1994. The pdhR-aceEF-lpd operon of Escherichia coli expresses the pyruvate dehydrogenase complex. Mol. Microbiol. 12:95-104. [DOI] [PubMed] [Google Scholar]
- 363.Rado, T. A., and J. A. Hoch. 1973. Phosphotransacetylase from Bacillus subtilis: purification and physiological studies. Biochim. Biophys. Acta 321:114-125. [DOI] [PubMed] [Google Scholar]
- 364.Ramakrishnan, R., M. Schuster, and R. B. Bourret. 1998. Acetylation of Lys-92 enhances signaling by the chemotaxis response regulator protein CheY. Proc. Natl. Acad. Sci. USA 95:4918-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Ramos, H. C., T. Hoffmann, M. Marino, H. Ndjari, E. Presecan-Siedel, O. Dreesen, P. Glaser, and D. Jahn. 2000. Fermentative metabolism of Bacillus subtilis: physiology and regulation of gene expression. J. Bacteriol. 182:3072-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Ramseier, T. M., S. Bledig, V. Michotey, R. Feghali, and M. H. Saier, Jr. 1995. The global regulatory protein FruR modulates the direction of carbon flow in Escherichia coli. Mol. Microbiol. 16:1157-1169. [DOI] [PubMed] [Google Scholar]
- 367.Ramseier, T. M., D. Negre, J. C. Cortay, M. Scarabel, A. J. Cozzone, and M. H. Saier, Jr. 1993. In vitro binding of the pleiotropic transcriptional regulatory protein, FruR, to the fru, pps, ace, pts and icd operons of Escherichia coli and Salmonella typhimurium. J. Mol. Biol. 234:28-44. [DOI] [PubMed] [Google Scholar]
- 368.Reeves, R. E., L. G. Warren, B. Susskind, and H. S. Lo. 1977. An energy-conserving pyruvate-to-acetate pathway in Entamoeba histolytica. Pyruvate synthase and a new acetate thiokinase. J. Biol. Chem. 252:726-731. [PubMed] [Google Scholar]
- 369.Reinscheid, D., S. Schnicke, D. Rittmann, U. Zahnow, H. Sahm, and B. Eikmanns. 1999. Cloning, sequence analysis, expression and inactivation of the Corynebacterium glutamicum pta-ack operon encoding phosphotransacetylase and acetate kinase. Microbiology 145:503-513. [DOI] [PubMed] [Google Scholar]
- 370.Reitzer, L. 2003. Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 57:155-176. [DOI] [PubMed] [Google Scholar]
- 371.Reitzer, L. J., and B. Magasanik. 1985. Expression of glnA in Escherichia coli is regulated at tandem promoters. Proc. Natl. Acad. Sci. USA 82:1979-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Reizer, J., J. Deutscher, and M. H. Saier, Jr. 1989. Metabolite-senstive, ATP- dependent, protein kinase catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system in Gram-positive bacteria. Biochemie 71:989-996. [DOI] [PubMed] [Google Scholar]
- 373.Reizer, J., C. Hoischen, F. Tigemeyer, C. Rivolta, R. Rabus, J. Stulke, D. Karamata, M. H. Saier, Jr., and W. Hillen. 1998. A novel protein kinase that controls carbon catabolite repression in bacteria. Mol. Microbiol. 27:1157-1169. [DOI] [PubMed] [Google Scholar]
- 374.Renna, M. C., N. Najimudin, L. R. Winik, and S. A. Zahler. 1993. Regulation of the Bacillus subtilis alsS, alsD, and alsR genes involved in post-exponential-phase production of acetoin. J. Bacteriol. 175:3863-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Repaske, D. R., and J. Adler. 1981. Change in intracellular pH of Escherichia coli mediates the chemotactic response to certain attractants and repellents. J. Bacteriol. 145:1196-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Reyrat, J. M., M. David, J. Batut, and P. Boistard. 1994. FixL of Rhizobium meliloti enhances the transcriptional activity of a mutant FixJD54N protein by phosphorylation of an alternate residue. J. Bacteriol. 176:1969-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Ricke, S. C. 2003. The gastrointestinal tract ecology of Salmonella enteritidis colonization in molting hens. Poult. Sci. 82:1003-1007. [DOI] [PubMed] [Google Scholar]
- 378.Rigden, D. J. 2003. Unexpected catalytic site variation in phosphoprotein phosphatase homologues of cofactor-dependent phosphoglycerate mutase. FEBS Lett. 536:77-84. [DOI] [PubMed] [Google Scholar]
- 379.Rinas, U., H. Kracke-Helm, and K. Schugerl. 1989. Glucose as a substrate in recombinant strain fermentation technology. Appl. Microbiol. Biotechnol. 31:163-167. [Google Scholar]
- 380.Riondet, C., R. Cachon, Y. Wache, G. Alcaraz, and C. Divies. 2000. Extracellular oxidoreduction potential modifies carbon and electron flow in Escherichia coli. J. Bacteriol. 182:620-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Roe, A. J., D. McLaggan, I. Davidson, C. O'Bryne, and I. R. Booth. 1998. Perturbation of anion balance during inhibition of growth of Escherichia coli by weak acids. J. Bacteriol. 180:767-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Roe, A. J., C. O'Byrne, D. McLaggan, and I. R. Booth. 2002. Inhibition of Escherichia coli growth by acetic acid: a problem with methionine biosynthesis and homocysteine toxicity. Microbiology 148:2215-2222. [DOI] [PubMed] [Google Scholar]
- 383.Roggiani, M., and D. Dubnau. 1993. ComA, a phosphorylated response regulator protein of Bacillus subtilis, binds to the promoter region of srfA. J. Bacteriol. 175:3182-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Romeo, T. 1998. Global regulation by the small RNA-binding protein CsrA and the non- coding RNA molecule CsrB. Mol. Microbiol. 29:1321-1330. [DOI] [PubMed] [Google Scholar]
- 385.Rose, I. A., M. Grunsberg-Manago, S. R. Korey, and S. Ochoa. 1954. Enzymatic phosphorylation of acetate. J. Biol. Chem. 211:737-756. [PubMed] [Google Scholar]
- 386.Rossman, R., G. Sawers, and A. Bock. 1991. Mechanism of regulation of the formate- hydrogenlyase pathway by oxygen, nitrate, and pH: definition of the formate regulon. Mol. Microbiol. 5:2807-2814. [DOI] [PubMed] [Google Scholar]
- 387.Ruff, J., K. Denger, and A. M. Cook. 2003. Sulphoacetaldehyde acetyltransferase yields acetyl phosphate: purification from Alcaligenes defragrans and gene clusters in taurine degradation. Biochem. J. 369:275-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Russell, J. B., and F. Diez-Gonzales. 1998. The effects of fermentation acids on bacterial growth. Adv. Microb. Physiol. 39:205-234. [DOI] [PubMed] [Google Scholar]
- 389.Saier, M. H., Jr., and T. M. Ramseier. 1996. The catabolite repressor/activator (Cra) protein of enteric bacteria. J. Bacteriol. 178:3411-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Salmond, C. V., R. G. Kroll, and I. R. Booth. 1984. The effect of food preservatives on pH homeostasis in Escherichia coli. J. Gen. Microbiol. 130:2845-2850. [DOI] [PubMed] [Google Scholar]
- 391.Sanchez, L. B., M. Y. Galperin, and M. Muller. 2000. Acetyl-CoA synthetase from the amitochondriate eukaryote Giardia lamblia belongs to the newly recognized superfamily of Acyl-CoA synthetases (nucleoside diphosphate-forming). J. Biol. Chem. 275:5794-5803. [DOI] [PubMed] [Google Scholar]
- 392.Sanchez, L. B., and M. Muller. 1996. Purification and characterization of the acetate forming enzyme, acetyl-CoA synthetase (ADP-forming) from the amitochondriate protist, Giardia lamblia. FEBS Lett. 378:240-244. [DOI] [PubMed] [Google Scholar]
- 393.Sanders, D. A., B. L. Gillece-Castro, A. L. Burlingame, and D. E. J. Koshland. 1992. Phosphorylation site of NtrC, a protein phosphatase whose covalent intermediate activates transcription. J. Bacteriol. 174:5117-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Sato, M., K. Machida, E. Arikado, H. Saito, T. Kakegawa, and H. Kobayashi. 2000. Expression of outer membrane proteins in Escherichia coli growing at acid pH. Appl. Environ. Microbiol. 66:943-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Schafer, T., and P. Schonheit. 1991. Pyruvate metabolism of the hyperthermophilic archaebacterium Pyrococcus furiosus. Acetate formation from acetyl-CoA and ATP synthesis are catalysed by an acetyl-CoA synthetase (ADP-forming). Arch. Microbiol. 155:366-377. [Google Scholar]
- 396.Schafer, T., M. Selig, and P. Schonheit. 1993. Acetyl-CoA synthetase (ADP-forming) in archaea, a novel enzyme involved in acetate and ATP synthesis. Arch. Microbiol. 159:72-83. [Google Scholar]
- 397.Scharrer, E., and T. Lutz. 1990. Effects of short chain fatty acids and K on absorption of Mg and other cations by the colon and caecum. Z. Ernahrungswiss. 29:162-168. [DOI] [PubMed] [Google Scholar]
- 398.Scheppach, W., E. W. Pomare, M. Elia, and J. H. Cummings. 1991. The contribution of the large intestine to blood acetate in man. Clin. Sci. (London) 80:177-182. [DOI] [PubMed] [Google Scholar]
- 399.Schmiel, D. H., G. M. Young, and V. L. Miller. 2000. The Yersinia enterocolitica phospholipase gene yplA is part of the flagellar regulon. J. Bacteriol. 182:2314-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Schonheit, P., and T. Schafer. 1995. Metabolism of hyperthermophiles. World J. Microbiol. Biotechnol. 11:26-57. [DOI] [PubMed] [Google Scholar]
- 401.Schrader, T., and J. R. Andreesen. 1992. Purification and characterization of protein PC, a component of glycine reductase from Eubacterium acidaminophilum. Eur. J. Biochem. 206:79-85. [DOI] [PubMed] [Google Scholar]
- 402.Sedewitz, B., K. H. Schleifer, and F. Gotz. 1984. Physiological role of pyruvate oxidase in the aerobic metabolism of Lactobacillus plantarum. J. Bacteriol. 160:462-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Sedewitz, B., K. H. Schleifer, and F. Gotz. 1984. Purification and biochemical characterization of pyruvate oxidase from Lactobacillus plantarum. J. Bacteriol. 160:273-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Semenza, G. 2001. Fifty years ago: the identification of ‘active acetate’ as acetyl-CoA. FEBS Lett. 509:343-344. [DOI] [PubMed] [Google Scholar]
- 405.Seufert, C. D., M. Graf, G. Janson, A. Kuhn, and H. D. Soling. 1974. Formation of free acetate by isolated perfused livers from normal, starved and diabetic rats. Biochem. Biophys. Res. Commun. 57:901-909. [DOI] [PubMed] [Google Scholar]
- 406.Shen, J., and R. P. Gunsalus. 1997. Role of multiple ArcA recognition sites in anaerobic regulation of succinate dehydrogenase (sdhCDAB) gene expression in Escherichia coli. Mol. Microbiol. 26:223-236. [DOI] [PubMed] [Google Scholar]
- 407.Shin, B.-S., S.-K. Choi, and S.-H. Park. 1999. Regulation of the Bacillus subtilis phosphotransacetylase gene. J. Biochem. (Tokyo) 126:333-339. [DOI] [PubMed] [Google Scholar]
- 408.Shin, S., and C. Park. 1995. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J. Bacteriol. 177:4696-4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Shin, S., J. Sheen, and C. Park. 1993. Suppression of high-temperature inhibition of motility due to the change in acetate metabolism in Escherichia coli K-12. Kor. J. Microbiol. 31:504-511. [Google Scholar]
- 410.Shin, S., S. G. Song, D. S. Lee, J. G. Pan, and C. Park. 1997. Involvement of iclR and rpoS in the induction of acs, the gene for acetyl coenzyme A synthetase of Escherichia coli K-12. FEMS Microbiol. Lett. 146:103-108. [DOI] [PubMed] [Google Scholar]
- 411.Silversmith, R. E., J. L. Appleby, and R. B. Bourret. 1997. Catalytic mechanism of phosphorylation and dephosphorylation of CheY: kinetic characterization of imidazole phosphates as phosphodonors and the role of acid catalysis. Biochemistry 36:14965-14974. [DOI] [PubMed] [Google Scholar]
- 412.Singh-Wissmann, K., and J. G. Ferry. 1995. Transcriptional regulation of the phosphotransacetylase-encoding and acetate kinase-encoding genes (pta and ack) from Methanosarcina thermophila. J. Bacteriol. 177:1699-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Smith, M. W., and F. C. Neidhardt. 1983. 2-Oxoacid dehydrogenase complexes of Escherichia coli: cellular amounts and patterns of synthesis. J. Bacteriol. 156:81-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Snoep, J. L., M. J. Teixeira de Mattos, P. W. Postma, and O. M. Niejssel. 1990. Involvement of pyruvate dehydrogenase in product formation in pyruvate-limited anaerobic chemostat cultures of Enterococcus faecalis NCTC 775. Arch. Microbiol. 154:50-55. [DOI] [PubMed] [Google Scholar]
- 415.Soler, F., M.-I. Fortea, A. Lax, and F. Fernandez-Belda. 2002. Dissecting the hydrolytic activities of sarcoplasmic reticulum ATPase in the presence of acetyl phosphate. J. Biol. Chem. 277:38127-38132. [DOI] [PubMed] [Google Scholar]
- 416.Sone, H., H. Shimano, Y. Sakakura, N. Inoue, M. Amemiya-Kudo, N. Yahagi, M. Osawa, H. Suzuki, T. Yokoo, A. Takahashi, K. Iida, H. Toyoshima, A. Iwama, and N. Yamada. 2002. Acetyl-coenzyme A synthetase is a lipogenic enzyme controlled by SREBP-1 and energy status. Am. J. Physiol. Endocrinol. Metab. 282:E222-E230. [DOI] [PubMed] [Google Scholar]
- 417.Sourjik, V., and R. Schmitt. 1998. Phosphotransfer between CheA, CheY1, and CheY2 in the chemotaxis signal transduction chain of Rhizobium meliloti. Biochemistry 37:2327-2335. [DOI] [PubMed] [Google Scholar]
- 418.Speck, E. L., and E. Freese. 1973. Control of metabolite secretion in Bacillus subtilis. J. Gen. Microbiol. 78:261-275. [DOI] [PubMed] [Google Scholar]
- 419.Spellerberg, B., D. R. Cundell, J. Sandros, B. J. Pearce, I. Idanpaan-Heikkila, C. Rosenow, and H. R. Masure. 1996. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol. Microbiol. 19:803-813. [DOI] [PubMed] [Google Scholar]
- 420.Spencer, M. E., and J. R. Guest. 1987. Regulation of citric acid cycle genes in facultative bacteria. Microbiol. Sci. 4:164-168. [PubMed] [Google Scholar]
- 421.Spencer, M. E., and J. R. Guest. 1985. Transcription analysis of the sucAB, aceEF, and lpd genes of Escherichia coli. Mol. Gen. Genet. 200:145-154. [DOI] [PubMed] [Google Scholar]
- 422.Stadtman, T., and J. Davis. 1991. Glycine reductase protein C. Properties and characterization of its role in the reductive cleavage of Se-carboxymethyl-selenoprotein A. J. Biol. Chem. 266:22147-22153. [PubMed] [Google Scholar]
- 423.Stadtman, T. C. 1989. Clostridial glycine reductase: protein C, the acetyl group acceptor, catalyzes the arsenate-dependent decomposition of acetyl phosphate. Proc. Natl. Acad. Sci. USA 86:7853-7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Stancik, L. M., D. M. Stancik, B. Schmidt, D. M. Barnhart, Y. N. Yoncheva, and J. L. Slonczewski. 2002. pH-Dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J. Bacteriol. 184:4246-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Starai, V., and J. Escalante-Semerena. 2004. Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J. Mol. Biol. 340:1005-1012. [DOI] [PubMed] [Google Scholar]
- 426.Starai, V. J., I. Celic, R. N. Cole, J. D. Boeke, and J. C. Escalante-Semerena. 2002. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298:2390-2392. [DOI] [PubMed] [Google Scholar]
- 427.Starai, V. J., and J. C. Escalante-Semerena. 2004. Acetyl-coenzyme A synthetase (AMP forming). Cell. Mol. Life Sci. 61:2020-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Starai, V. J., H. Takahashi, J. D. Boeke, and J. C. Escalante-Semerena. 2003. Short- chain fatty acid activation by acyl-coenzyme A synthetases requires SIR2 protein function in Salmonella enterica and Saccharomyces cerevisiae. Genetics 163:545-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Steeg, P. S., D. Palmieri, T. Ouatas, and M. Salerno. 2003. Histidine kinases and histidine phosphorylated proteins in mammalian cell biology, signal transduction and cancer. Cancer Lett. 190:1-12. [DOI] [PubMed] [Google Scholar]
- 430.Sterri, S. H., and F. Fonnum. 1980. Acetyl-CoA synthesizing enzymes in cholinergic nerve terminals. J. Neurochem. 35:249-254. [DOI] [PubMed] [Google Scholar]
- 431.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 432.Stokes, J. L. 1949. Fermentation of glucose by suspensions of Escherichia coli. J. Bacteriol. 57:147-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Stout, V. 1994. Regulation of capsule synthesis includes interactions of the RcsC/RcsB regulatory pair. Res. Microbiol. 145:389-392. [DOI] [PubMed] [Google Scholar]
- 434.Stulke, J., and W. Hillen. 1999. Carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 2:195-201. [DOI] [PubMed] [Google Scholar]
- 435.Summers, M. L., M. C. Denton, and T. R. McDermott. 1999. Genes coding for phosphotransacetylase and acetate kinase in Sinorhizobium meliloti are in an operon that is inducible by phosphate stress and controlled by phoB. J. Bacteriol. 181:2217-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Suzuki, K., X. Wang, T. Weilbacher, A.-K. Pernestig, O. Melefors, D. Georgellis, P. Babitzke, and T. Romeo. 2002. Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J. Bacteriol. 184:5130-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Suzuki, T. 1969. Phosphotransacetylase of Escherichia coli B, activation by pyruvate and inhibition by NADH and certain nucleotides. Biochim. Biophys. Acta 191:559-569. [DOI] [PubMed] [Google Scholar]
- 438.Takamura, Y., and G. Nomura. 1988. Changes in the intracellular concentration of acetyl-CoA and malonyl-CoA in relation to the carbon and energy metabolism of Escherichia coli K12. J. Gen. Microbiol. 134:2249-2253. [DOI] [PubMed] [Google Scholar]
- 439.Takeda, S., A. Matsushika, and T. Mizuno. 1999. Repression of the gene encoding succinate dehydrogenase in response to glucose is mediated by the EIICB(Glc) protein in Escherichia coli. J. Biochem. (Tokyo) 126:354-360. [DOI] [PubMed] [Google Scholar]
- 440.Tao, H., C. Bausch, C. Richmond, F. R. Blattner, and T. Conway. 1999. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 181:6425-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Thauer, R. K., K. Jungermann, and K. Decker. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41:100-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Thomason, P. A., D. Traynor, J. B. Stock, and R. R. Kay. 1999. The RdeA-RegA system, a eukaryotic phospho-relay controlling cAMP breakdown. J. Biol. Chem. 274:27379-27384. [DOI] [PubMed] [Google Scholar]
- 443.Thomason, P. A., D. Traynor, G. Cavet, W. T. Chang, A. J. Harwood, and R. R. Kay. 1998. An intersection of the cAMP/PKA and two-component signal transduction systems of Dictyostelium. EMBO J. 17:2838-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Tittmann, K., D. Proske, M. Spinka, S. Ghisla, R. Rudolph, G. Hubner, and G. Kern. 1998. Activation of thiamin diphosphate and FAD in the phosphate dependent pyruvate oxidase from Lactobacillus plantarum. J. Biol. Chem. 273:12929-12934. [DOI] [PubMed] [Google Scholar]
- 445.Toh, H. 1990. N-terminal halves of gramicidin S synthetase 1, and tyrocidine synthetase 1 as novel members of firefly luciferase family. Protein Seq. Data Anal. 3:517-521. [PubMed] [Google Scholar]
- 446.Toh, H. 1991. Sequence analysis of firefly luciferase family reveals a conservative sequence motif. Protein Seq. Data Anal. 4:111-117. [PubMed] [Google Scholar]
- 447.Topping, D. L., and P. M. Clifton. 2001. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 81:1031-1064. [DOI] [PubMed] [Google Scholar]
- 448.Tran, V. K., R. Oropeza, and L. J. Kenney. 1999. A single amino acid substitution in the C terminus of OmpR alters DNA recognition and phosphorylation. J. Mol. Biol. 299:1257-1270. [DOI] [PubMed] [Google Scholar]
- 449.Tsang, A., and J. Escalante-Semerena. 1996. cobB function is required for catabolism of propionate in Salmonella typhimurium LT2: evidence for existence of a substitute function for CobB within the 1,2-propanediol utilization (pdu) operon. J. Bacteriol. 178:7016-7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Tsang, A. W., A. R. Horswill, and J. C. Escalante-Semerena. 1998. Studies of regulation of expression of the propionate (prpBCDE) operon provide insights into how Salmonella typhimurium LT2 integrates its 1,2-propanediol and propionate catabolic pathways. J. Bacteriol. 180:6511-6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Tseng, G. C., M.-K. Oh, L. Rohlin, J. C. Liao, and W. H. Wong. 2001. Issues in cDNA microarray analysis: quality filtering, channel normalization, models of variations and assessment of gene effects. Nucleic Acids. Res. 29:2549-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Turinsky, A. J., F. J. Grundy, J.-H. Kim, G. H. Chambliss, and T. M. Henkin. 1998. Transcriptional activation of the Bacillus subtilis ackA gene requires sequences upstream of the promoter. J. Bacteriol. 180:5961-5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Turinsky, A. J., T. R. Moir-Blais, F. J. Grundy, and T. M. Henkin. 2000. Bacillus subtilis ccpA gene mutants specifically defective in activation of acetoin biosynthesis. J. Bacteriol. 182:5611-5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Vallari, D. S., and S. Jackowski. 1988. Biosynthesis and degradation both contribute to the regulation of coenzyme A content in Escherichia coli. J. Bacteriol. 170:3961-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.van den Berg, M. A., P. de Jong-Gubbels, C. J. Kortland, J. P. van Dijken, J. T. Pronk, and H. Y. Steensma. 1996. The two acetyl-coenzyme A synthetases of Saccharomyces cerevisiae differ with respect to kinetic properties and transcriptional regulation. J. Biol. Chem. 271:28953-28959. [DOI] [PubMed] [Google Scholar]
- 456.van de Walle, M., and J. Shiloach. 1998. Proposed mechanism of acetate accumulation in two recombinant Escherichia coli strains during high density fermentation. Biotechnol. Bioeng. 57:71-78. [DOI] [PubMed] [Google Scholar]
- 457.Van Dyk, T. K., and R. A. LaRossa. 1987. Involvement of ack-pta operon products in alpha-ketobutyrate metabolism by Salmonella typhimurium. Mol. Gen. Genet. 207:435-440. [DOI] [PubMed] [Google Scholar]
- 458.Varma, A., and B. O. Paulson. 1994. Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Appl. Environ. Microbiol. 60:3724-3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Voet, D., and J. G. Voet. 1990. Biochemistry, John Wiley & Sons, Inc., New York, N.Y.
- 460.Wadolkowski, E. A., D. C. Laux, and P. S. Cohen. 1988. Colonization of the streptomycin-treated mouse large intestine by a human fecal Escherichia coli strain: role of growth in mucus. Infect. Immun. 56:1030-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Wagner, A., S. Schultz, J. Bomke, T. Pils, W. Lehmann, and J. Knappe. 2001. YfiD of Escherichia coli and Y061 of bacteriophage T4 as autonomous glycyl radical cofactors reconstituting the catalytic center of oxygen-fragmented pyruvate formate-lyase. Biochem. Biophys. Res. Commun. 285:456-462. [DOI] [PubMed] [Google Scholar]
- 462.Wagner, M., D. Sonntag, R. Grimm, A. Pich, C. Eckerskorn, B. Sohling, and J. R. Andreesen. 1999. Substrate-specific selenoprotein B of glycine reductase from Eubacterium acidaminophilum. Biochemical and molecular analysis. Eur. J. Biochem. 260:38-49. [DOI] [PubMed] [Google Scholar]
- 463.Wanner, B. L. 1993. Gene regulation by phosphate in enteric bacteria. J. Cell. Biochem. 51:47-54. [DOI] [PubMed] [Google Scholar]
- 464.Wanner, B. L. 1992. Is cross regulation by phosphorylation of two-component response regulator proteins important in bacteria? EMBO J. 11:265-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Wanner, B. L. 1996. Phosphorus assimilation and control of the phosphate regulon, p. 1357-1381. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 466.Wanner, B. L., and M. R. Wilmes-Riesenberg. 1992. Involvement of phosphotransacetylase, acetate kinase, and acetyl phosphate synthesis in control of the phosphate regulon in Escherichia coli. J. Bacteriol. 174:2124-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Wei, B. L., A.-M. Brun-Zinkernagel, J. W. Simecka, B. M. Pruss, P. Babitzke, and T. Romeo. 2001. Positive regulation of motility and flhDC expression by the RNA- binding protein CsrA of Escherichia coli. Mol. Microbiol. 40:245-256. [DOI] [PubMed] [Google Scholar]
- 468.Weickert, M. J., and G. H. Chambliss. 1990. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilits. Proc. Natl. Acad. Sci. USA 87:6238-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Wendisch, V. F. 2003. Genome-wide expression analysis in Corynebacterium glutamicum using DNA microarrays. J. Biotechnol. 104:273-285. [DOI] [PubMed] [Google Scholar]
- 470.Wendisch, V. F., M. Spies, D. J. Reinscheid, S. Schnicke, H. Sahm, and B. J. Eikmanns. 1997. Regulation of acetate metabolism in Corynebacterium glutamicum: transcriptional control of the isocitrate lyase and malate synthase genes. Arch. Microbiol. 168:262-269. [DOI] [PubMed] [Google Scholar]
- 471.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26:369-376. [DOI] [PubMed] [Google Scholar]
- 472.Wilde, R. J., and J. R. Guest. 1986. Transcript analysis of the citrate synthase and succinate dehydrogenase genes of Escherichia coli K12. J. Gen. Microbiol. 132:3239-3251. [DOI] [PubMed] [Google Scholar]
- 473.Wolfe, A. J., D.-E. Chang, J. D. Walker, J. E. Seitz-Partridge, M. D. Vidaurri, C. F. Lange, B. M. Prüß, M. C. Henk, J. C. Larkin, and T. Conway. 2003. Evidence that acetyl phosphate functions as a global signal during biofilm development. Mol. Microbiol. 48:977-988. [DOI] [PubMed] [Google Scholar]
- 474.Wolfe, A. J., M. P. Conley, and H. C. Berg. 1988. Acetyladenylate plays a role in controlling the direction of flagellar rotation. Proc. Natl. Acad. Sci. USA 85:6711-6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Woodnutt, G., and D. S. Parker. 1986. Acetate metabolism by tissues of the rabbit. Comp. Biochem. Physiol. Ser. B. 85:487-490. [DOI] [PubMed] [Google Scholar]
- 476.Wyborn, N. R., S. L. Messenger, R. A. Henderson, G. Sawers, R. E. Roberts, M. M. Attwood, and J. Green. 2002. Expression of the Escherichia coli yfiD gene responds to intracellular pH and reduces the accumulation of acidic metabolic end products. Microbiology 148:1015-1026. [DOI] [PubMed] [Google Scholar]
- 477.Xu, B., M. Jahic, and S.-O. Enfors. 1999. Modeling of overflow metabolism in batch and fed-batch cultures of Escherichia coli. Biotechnol. Prog. 15:81-90. [DOI] [PubMed] [Google Scholar]
- 478.Yamamoto, K., and A. Ishihama. 2003. Two different modes of transcription repression of the Escherichia coli acetate operon by IcIR. Mol. Microbiol. 47:183-194. [DOI] [PubMed] [Google Scholar]
- 479.Yamamoto-Otake, H. M., A. Matsuyama, and F. Nakano. 1990. Cloning of a gene coding for phosphotransacetylase from Escherichia coli. Appl. Microbiol. Biotechnol. 33:680-682. [DOI] [PubMed] [Google Scholar]
- 480.Yamashita, H., A. Fukuura, T. Nakamura, T. Kaneyuki, M. Kimoto, M. Hiemori, and H. Tsuji. 2002. Purification and partial characterization of acetyl-coA synthetase in rat liver mitochondria. J. Nutr. Sci. Vitaminol. (Tokyo) 48:359-364. [DOI] [PubMed] [Google Scholar]
- 481.Yamashita, H., T. Kaneyuki, and K. Tagawa. 2001. Production of acetate in the liver and its utilization in peripheral tissues. Biochim. Biophys. Acta 1532:79-87. [DOI] [PubMed] [Google Scholar]
- 482.Yang, Y. T., A. A. Aristidou, K. Y. San, and G. N. Bennett. 1999. Metabolic flux analysis of Escherichia coli deficient in the acetate production pathway and expressing the Bacillus subtilis acetolactate synthase. Metab. Eng. 1:26-34. [DOI] [PubMed] [Google Scholar]
- 483.Yang, Y. T., G. N. Bennett, and K. Y. San. 1999. Effect of inactivation of nuo and ackA-pta on redistribution of metabolic fluxes in Escherichia coli. Biotechnol. Bioeng. 65:291-297. [PubMed] [Google Scholar]
- 484.Zahrt, T. C., C. Wozniak, D. Jones, and A. Trevett. 2003. Functional analysis of the Mycobacterium tuberculosis MprAB two-component signal transduction system. Infect. Immun. 71:6962-6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Zalieckas, J. M., J. Wray, L. V., and S. H. Fisher. 1998. Expression of the Bacillus subtilis acsA gene: position and sequence context affect cre-mediated carbon catabolite repression. J. Bacteriol. 180:6649-6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Zapf, J. W., J. A. Hoch, and J. M. Whiteley. 1996. A phosphotransferase activity of the Bacillus subtilis sporulation protein Spo0F that employs phosphoramidate substrates. Biochemistry 35:2926-2933. [DOI] [PubMed] [Google Scholar]
- 487.Zeeman, A. M., and H. Y. Steensma. 2003. The acetyl co-enzyme A synthetase genes of Kluyveromyces lactis. Yeast 20:13-23. [DOI] [PubMed] [Google Scholar]
- 488.Zhu, P. P., and A. Peterkofsky. 1996. Sequence and organization of genes encoding enzymes involved in pyruvate metabolism in Mycoplasma capricolum. Protein Sci. 5:1719-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]