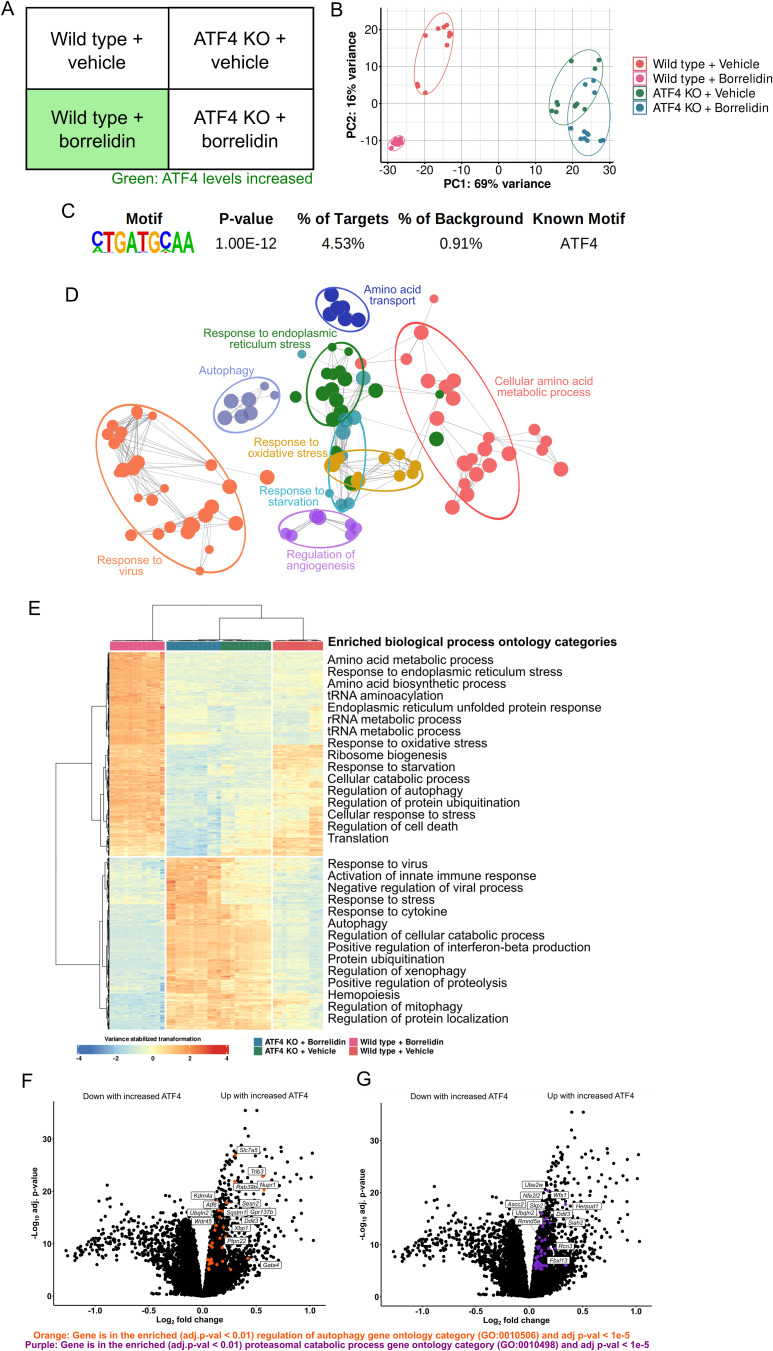
Fig. 3.
ATF4 causes the differential expression of genes involved with protein turnover. A RNASeqblock design to control for borrelidin treatment and Atf4 knockout sequencing. B Principal component analysis of the 46 samples sequenced. C HOMER motif enrichment from the differentially expressed genes associated with ATF4 (adj. p-value < 1e-10). D) ClueGo enriched biological process ontology categories (adj. p-value < 0.01) of differentially expressed genes (adj. p-value < 1e-10) from the linear model result (Design = ~ ATF4 + Condition). E Heatmap and hierarchical clustering of linear model results (p adj < 1e-10). Biological process gene ontology enrichment utilized Panther’s ontology resource. F, G Volcano plots of genes differentially expressed from linear model. Both regulation of autophagy (in orange) and proteasomal protein catabolic process (in purple) were found to be enriched in the genes upregulated (p adj < 1e-10) with ATF4 (Panther’s ontology resource, adj. p-value < 0.01). “Up with increased ATF4” and “Down with increased ATF4” are labeled on either side of the volcano plots presented in association with the limma linear model fit to ATF4 activity given by the linear model study design (Design = ~ ATF4 + Genotype)