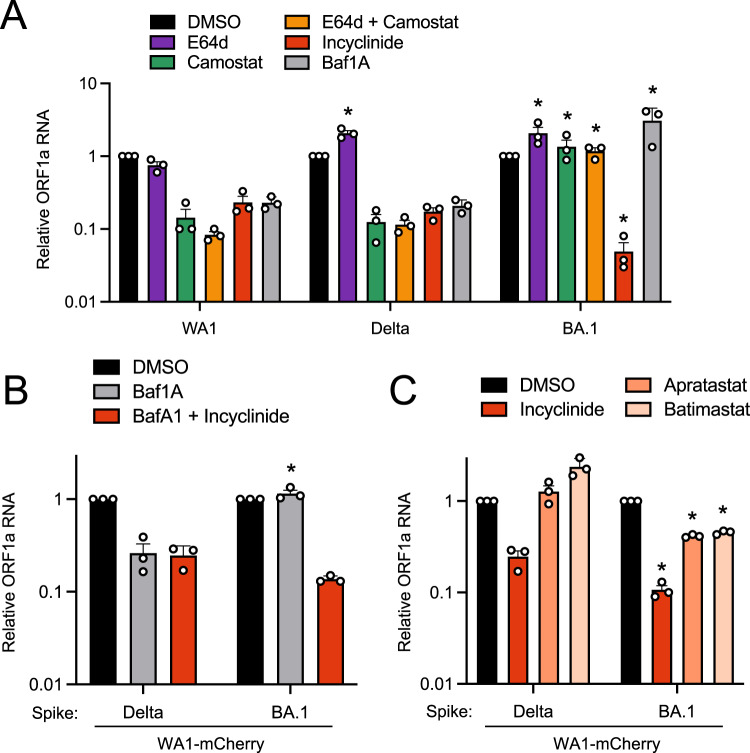
Fig. 3. Entry of Omicron BA.1 into primary nasal epithelial cells is sensitive to MMP inhibitors but not inhibitors of cathepsins or TMPRSS2.
A Primary human nasal epithelial cells (pooled from 14 donors) were cultured at the air-liquid interface and pretreated with 10 µM E64d, 10 µM camostat mesylate, a combination of 10 µM E64d and 10 µM camostat mesylate, 10 µM incyclinide, or 1 µM bafilomycin A1 for 2 h. Cells were challenged with WA.1, Delta, or BA.1 at an MOI of 0.05. At 48 h post-inoculation, RNA was extracted from cells, and ORF1a levels were measured by RT-qPCR. For each virus, ORF1a abundance in the DMSO condition was set to 1. Results are represented as means plus standard error from three independent infections (biological replicates). Statistically significant differences (*P < 0.05) between the indicated condition and the corresponding condition of WA.1 were determined by one-way ANOVA adjusted for multiple comparisons (exact p values from left to right: 0.0025, 0.0372, 0.019, 0.0013, 0.0236, 0.0313). B Primary human nasal epithelial cells were cultured at the air–liquid interface and pretreated with 1 µM bafilomycin A1 or 1 µM bafilomycin A1 and 10 µM incyclinide for 2 h. Cells were challenged with WA1 (Delta Spike) or WA1 (BA.1 Spike) at an MOI of 0.05. At 48 h post-inoculation, ORF1a levels were measured by RT-qPCR. For each virus, ORF1a abundance in the DMSO condition was set to 1. Statistically significant differences (*P < 0.05) between the indicated condition and the corresponding condition of WA.1 (Delta Spike) were determined by the student’s unpaired two-sided t test (exact p value: 0.0015). C As in B, except primary human nasal epithelial cells cultured at the air–liquid interface were pretreated with 10 µM incyclinide, 10 µM apratastat, or 10 µM batimastat for 2 h and challenged with WA1 (Delta Spike) or WA1 (BA.1 Spike). Statistically significant differences (*P < 0.05) between the indicated condition and the corresponding condition of WA1 (Delta Spike) were determined by the student’s unpaired two-sided t test (exact p values from left to right: 0.0254, 0.0121, 0.0038). All results are represented as means plus standard error from three independent infections (symbols represent biological replicates). Refer to Supporting Dataset 1 for non-normalized data. Source data are provided as a Source Data file.