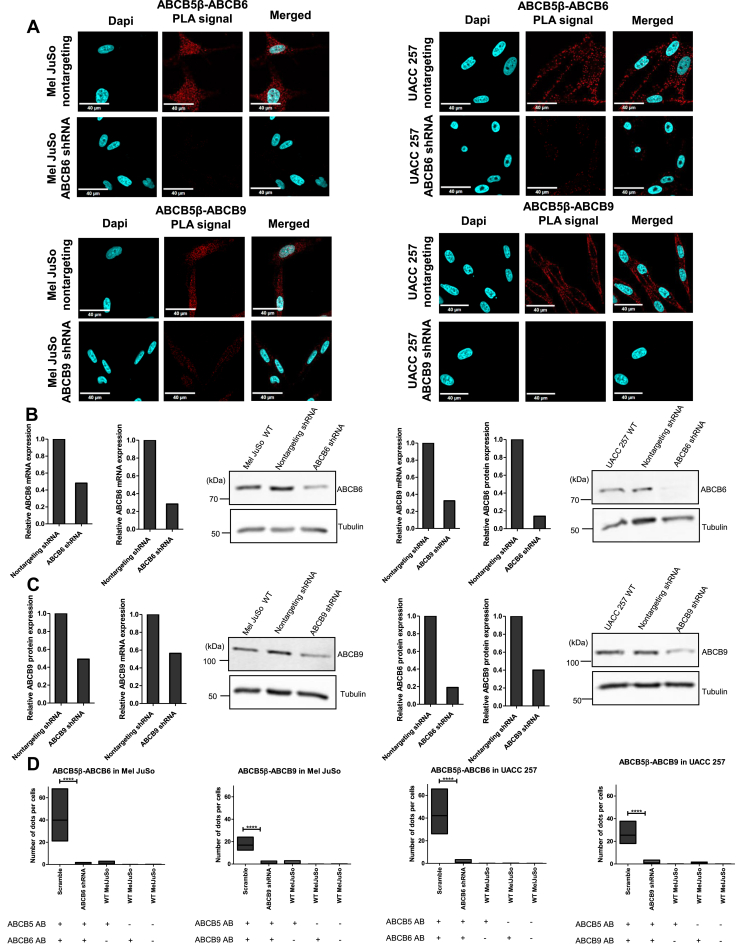
Figure 4.
Proximity ligation assay confirms the interactions between ABCB5β-ABCB6 and ABCB5β-ABCB9 protein pairs in Mel JuSo and UACC-257.A, confocal images of PLA in Mel JuSo and UACC-257 that stably expressed either a nontargeting shRNA or an ABCB6-specific (or ABCB9-specific) shRNA. The detected PLA signal is in red. Nuclei are stained in blue using DAPI. The scale bar represents 40 μM. After shRNA knockdown, efficiency was determined using reverse transcription-quantitative polymerase chain reaction and Western blots in Mel JuSo (B) and UACC-257 (C). 18S and tubulin were used as references for reverse transcription-quantitative polymerase chain reaction and Western blots normalization, respectively (n = 2). ImageJ (https://imagej.net/ij/download.html) was used to quantify protein expression in the Western blot as well as the average number of dots per cell (n = 3) (D). Scramble and shRNA quantifications are shown along with technical negative controls, including PLA conducted using either one antibody (AB) or none. Data is presented as mean ± SD. In Mel JuSo: ABCB6 scramble (39.89 ± 14.21), ABCB6 shRNA (0.28 ± 0.60), ABCB5 AB control (0.55 ± 0.91), ABCB6 AB control (0 ± 0), ABCB9 scramble (16.89 ± 4.27), ABCB9 shRNA (0.84 ± 1.04), ABCB9 AB control (0 ± 0.01), and no antibody control (0.01 ± 0.05). In UACC-257: ABCB6 scramble (42.16 ± 10.62), ABCB6 shRNA (0.97 ± 1.00), ABCB5 AB control (0.01 ± 0.02), ABCB6 AB control (0.03 ± 0.07), ABCB9 scramble (25.39 ± 6.74), ABCB9 shRNA (1.04 ± 1.16), ABCB9 AB control (0.79 ± 0.58), and no antibody control (0 ± 0). Student’s t test was performed between the scramble and shRNA conditions. Statistical significances are presented on graph as ∗∗∗∗p < 0.0001. ABCB5β/B6 in Mel JuSo (t = 9.648, df = 22), ABCB5β/B9 in Mel JuSo (t = 12,66, df = 22), ABCB5β/B6 in UACC-257 (t = 13,38, df = 22), ABCB5β/B9 in UACC-257 (t = 12,34, df = 22). DAPI, 4′,6-diamidino-2-phenylindole.