Abstract
Salmonella isolates (n = 209) obtained from food animals and foods in Alberta during 1996 through 1999 were tested for sensitivity to 17 antimicrobials. Of the 3553 antimicrobial susceptibility tests on Salmonella isolates, 11.8% were positive for resistance. These isolates were commonly resistant to tetracycline (35.4%), streptomycin (32.5%), sulfamethoxazole (28.7%), ticarcillin (27.3%), and ampicillin (26.8%). Resistance to at least 1 antimicrobial was observed in 112 isolates (53.6%). Salmonella Typhimurium, S. Typhimurium var. Copenhagen, and S. Heidelberg were the most common serovars among isolates resistant to individual antimicrobials and multiple antimicrobials. The most common profile of multiple-antimicrobial resistance was that which included resistance to ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, tetracycline, and ticarcillin. The proportions of isolates that were resistant to antimicrobials were greater among bovine isolates of Salmonella than among poultry isolates, and this difference was greater among isolates from veterinary diagnostic sources than among those from monitoring sources.
Abstract
Résumé — Résistance antimicrobienne d’un choix d’isolats de Salmonella provenant d’animaux de boucherie et de denrées alimentaires en Alberta. En Alberta, entre 1996 et 1999, des isolats de Salmonella (n = 209) obtenus à partir d’animaux de boucherie et de denrées alimentaires ont été testés pour leur sensibilité à 17 antimicrobiens. Sur les 3553 tests de sensibilité des isolats de Salmonella aux antimicrobiens, 11,8 % montraient de la résistance. Ces isolats étaient fréquemment résistants à la tétracycline (35,4 %), à la streptomycine (32,5 %), au sulfaméthoxazole (28,7 %), à la ticarcilline (27,3 %) et à l’ampicilline (26,8 %). De la résistance à au moins 1 antimicrobien a été observée dans 112 isolats (53,6 %). Salmonelle typhymurium, S. typhymurium var Copenhagen et S. heidelberg étaient les sérovars les plus communs parmi les isolats résistant à un ou plusieurs anti-microbiens. Le profil le plus répandu de résistance multiple était celui comprenant une résistance à l’ampicilline, au chloramphénicol, à la streptomycine, au sulfaméthoxazole, à la tétracycline et à la ticarcilline. La proportion des isolats résistants aux antimicrobiens était plus élevée chez les isolats bovins de Salmonella que chez les isolats aviaires et cette différence était plus marquée parmi les isolats provenant de sources diagnostiques vétérinaires que parmi ceux de sources de contrôle sanitaire.
(Traduit par Docteur André Blouin)
Introduction
Antimicrobial resistance in pathogenic bacteria of animal and human origin is a major public health issue. Epidemiological and molecular methods have been used to suggest that antimicrobial use in animal agriculture and antimicrobial resistant bacteria from food animals can lead to antimicrobial resistant Salmonella, Campylobacter, and Enterococcus infections in humans (1–4). Continuous monitoring and closed surveys of antimicrobial resistance have been established in many countries to assess the impact on public health of antimicrobial resistance among bacteria in food animals and foods. Antimicrobial resistance in Salmonella is used in surveillance systems in the European Union and United States as an indicator of the status of resistance in zoonotic bacteria (5–7). Recently, the Office International des Épizooties (OIE) (World Organization for Animal Health) has initiated the development of international recommendations on the detection and control of antimicrobial resistance as it relates to zoonotic bacteria and to resistance determinants that may be transferred between animals and from animals to humans (8). Surveillance information is necessary to determine the proportion of resistance to antimicrobials in defined populations, detect emerging resistance trends, provide a basis for policy recommendations and interventions within the animal and public health fields, assess the impact of interventions, and provide information for prescribing practices and prudent use recommendations (8). In 1999, in response to increasing concerns regarding the emergence and spread of antimicrobial resistance worldwide, the Food Safety Division of Alberta Agriculture, Food and Rural Development tested 209 Salmonella isolates obtained from food animals and food in Alberta through veterinary diagnostic and monitoring systems from 1996 through 1999. The purpose of this study was to evaluate antimicrobial resistance and resistance profiles of Salmonella strains isolated from poultry, cattle, and pigs, and foods of animal origin in Alberta.
Materials and methods
Selection of isolates
Isolates (n = 209) were purposely selected by the Agri-Food Laboratories Branch, Food Safety Division of Alberta Agriculture, Food and Rural Development. The Salmonella isolates were from food animals or foods obtained through monitoring programmes or as voluntary diagnostic isolates submitted from 1996 through 1999. The isolates were selected to represent the Salmonella serovar distribution of all isolates collected during this period.
Source of isolates and serovar distribution
Salmonella were isolated at the Agri-Food Laboratories Branch, Food Safety Division of Alberta Agriculture, Food and Rural Development. Standard protocols for the isolation of Salmonella from fecal, environmental, and other sources were used (9). Isolates were transferred on Columbia slants to the Health Canada Salmonella Typing Laboratory of the Laboratory for Foodborne Zoonoses (an OIE Reference Laboratory for Salmonellosis) for serotyping and phage typing.
Testing for antimicrobial sensitivity
Each of the isolates was tested for susceptibility to a panel of 17 antimicrobials, including ampicillin, ticarcillin, amoxicillin-clavulanic acid, ceftiofur, ceftriaxone, cephalothin, amikacin, apramycin, gentamicin, kanamycin, streptomycin, nalidixic acid, ciprofloxacin, chloramphenicol, sulfamethoxazole, trimethoprim-sulfamethoxazole, and tetracycline, by using antimicrobial susceptibility plates in an automated system (Sensititre; TREK Diagnostic Systems, Cleveland, Ohio, USA). Antimicrobial sensitivity of isolates was evaluated according to classification guidelines suggested by the National Committee for Clinical Laboratory Standards (NCCLS) for humans (10) or animals (11), or the National Antimicrobial Resistance Monitoring System (12). When the minimum inhibitory concentration (MIC) of a Salmonella isolate for a given antimicrobial was in the intermediate-sensitivity classification for that antimicrobial, it was considered to be not resistant to that antimicrobial.
Data analysis
The lowest concentrations of antimicrobial that completely inhibit the growth of 50% and 90% of the isolates are represented by MIC50 and MIC90, respectively. The antimicrobial resistance of a group of isolates was calculated as the percentage of isolates among the group that were resistant to a single antimicrobial or a number of antimicrobials. Resistance was also evaluated in terms of percentage resistance (13), in which the denominator is the number of antimicrobial resistance tests conducted on isolates within a group. For this study, percentage resistance measures the resistance among a group of isolates averaged over the 17 antimicrobials. Percentage resistance was calculated as:
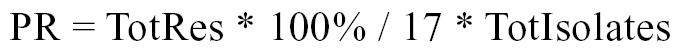 |
Where PR = percentage resistance for a group of isolates; TotRes = the number of antimicrobials to which each isolate within a group was resistant, summed over the number of isolates in the group; and TotIsolates = number of isolates tested within a group.
The association between the animal source of Salmonella isolates and resistance to antimicrobials was tested by calculating odds ratios and asymptotic 95% confidence intervals for the odds ratios (Statistical Package for the Social Sciences [SPSS], version 10.0; SPSS, Chicago, Illinois, USA). Poultry was the reference category against which bovine isolates were contrasted. Swine isolates were not included due to the small sample size. The data were further stratified by submission source, and stratified odds ratios for the association between animal source and antimicrobial resistance were calculated. Breslow-Day tests were used to assess homogeneity among stratified odds ratios. Mantel-Haenszel odds ratios were calculated when there was no statistical evidence against homogeneity across the strata. Similar analyses were conducted to assess the difference in antimicrobial resistance between isolates from monitoring and veterinary diagnostic sources while adjusting for animal source.
Results
Sources of isolates included poultry production facilities, food processors, veterinary clinics, and a Salmonella outbreak investigation; the numbers of isolates from these sources were 98, 43, 64, and 4, respectively. The majority of the isolates tested in this study were obtained from active monitoring sources and the majority of those originated from poultry (Table 1). Salmonella isolates from bovine sources were from predominantly diagnostic submissions. Among 209 Salmonella isolates, 51.2% belonged to serogroup B. Salmonella Typhimurium (35.3%), S. Heidelberg (20.6%), S. Typhimurium var. Copenhagen (14.7%), and S. Muenster (10.3%) were the most common serovars among veterinary diagnostic isolates. Salmonella Heidelberg (21.3%), S. Typhimurium (12.8%), S. Mbandaka (9.9%), S. Hadar (9.9%), and S. Kentucky (9.9%) were the most common serovars among monitoring isolates.
Table 1.
Distribution of Salmonella isolates among animal sources and submission sources that were screened for antimicrobial resistance
| Type of isolate | |||
|---|---|---|---|
| Animal source | Active monitoring | Diagnostic | Total |
| Poultry | 126 | 28 | 154 |
| Bovine | 8 | 34 | 42 |
| Swine | 7 | 6 | 13 |
| Total | 141 | 68 | 209 |
Half of the Salmonella isolates had MICs below the breakpoints for resistance to the 17 antimicrobials (Table 2). The range of MICs for ampicillin, ticarcillin, streptomycin, sulfamethoxazole, and tetracycline was wide and resistance to these antimicrobials was commonly observed. Among all antimicrobial groups, the quinolone antimicrobials, nalidixic acid and ciprofloxacin, had the greatest activity against the Salmonella isolates. Cross-resistance to ampicillin and ticarcillin was evident; 56 of the 57 isolates that were resistant to ticarcillin were also resistant to ampicillin.
Table 2.
Antimicrobial resistance and antimicrobial sensitivity of Salmonella isolates (n = 209)
| Antimicrobial | Resistancea (%) | MIC50 (μg/mL) | MIC90 (μg/mL) |
|---|---|---|---|
| Ampicillin | 26.8 | 2 | > 64 |
| Ticarcillin | 27.3 | 4 | > 128 |
| Amoxicillin-clavulanic acid | 9.6 | 1/0.5 | 16/8 |
| Cephalothin | 8.1 | 4 | 16 |
| Ceftiofur | 1.0 | 0.5 | 1 |
| Ceftriaxone | 0.0 | 0.25 | 0.25 |
| Amikacin | 0.0 | 4 | 4 |
| Apramycin | 0.0 | 2 | 4 |
| Gentamicin | 7.7 | 0.5 | 1 |
| Kanamycin | 9.6 | 16 | 16 |
| Streptomycin | 32.5 | 32 | 128 |
| Nalidixic acid | 0.5 | 4 | 4 |
| Ciprofloxacin | 0.0 | 0.015 | 0.015 |
| Sulfamethoxazole | 28.7 | 128 | > 512 |
| Trimethoprim-sulfamethoxazole | 1.0 | 0.12/2.38 | 0.25/4.75 |
| Tetracycline | 35.4 | 4 | > 64 |
| Chloramphenicol | 12.9 | 8 | > 32 |
a Breakpoints for resistance (μg/mL): ampicillin, ≥32; ticarcillin, ≥128; amoxicillin-clavulanic acid, ≥32/16; cephalothin, ≥32; ceftiofur, ≥8; ceftriaxone, ≥64; amikacin, ≥64; apramycin, ≥32; gentamicin, ≥16; kanamycin, ≥64; streptomycin, ≥64; nalidixic acid, ≥32; ciprofloxacin, ≥4; sulfamethoxazole, ≥512; trimethoprim-sulfamethoxazole, ≥4/76; tetracycline, ≥16; chloramphenicol, ≥32
MIC — Minimum inhibitory concentration
Among the 3553 antimicrobial-resistance tests performed on the 209 Salmonella isolates, 419 (11.8%) were positive for resistance (Table 3). Overall, 112 of the isolates tested (53.6%) were resistant to at least 1 antimicrobial. Among strains belonging to serogroup B, the proportion that were resistant to ampicillin, ticarcillin, streptomycin, and sulfamethoxazole ranged between 42% and 51%, while resistance to these drugs among strains of serogroups C, D, and E was much lower and ranged between 0% and 32% (data not shown). Percentage resistance to all antimicrobials was greatest among strains of S. Typhimurium, S. Typhimurium var. Copenhagen, S. Derby, and S. Heidelberg (Table 3). Among S. Typhimurium and S. Typhimurium var. Copenhagen (n = 54), resistance to ampicillin, ticarcillin, sulfamethoxazole, streptomycin, and tetracycline, singly, ranged between 61% and 69%. Among S. Heidelberg (n = 44), resistance to these antimicrobials ranged between 13% and 41%.
Table 3.
Antimicrobial resistance among Salmonella isolates within serogroups and serovars
| Serogroup or serovar | Number of isolates | Percentage resistancea | Resistance to ≥1 antimicrobial (%) |
|---|---|---|---|
| B | 107 | 18.9 | 69.2 |
| Heidelberg | 44 | 14.6 | 65.9 |
| Typhimurium | 42 | 23.0 | 69.0 |
| Typhimurium var. Copenhagen | 12 | 30.9 | 91.7 |
| Agona | 4 | 7.4 | 100.0 |
| Schwarzengrund | 3 | 0.0 | 0.0 |
| Derby | 1 | 17.7 | 100.0 |
| Reading | 1 | 0.0 | 0.0 |
| C | 77 | 4.7 | 40.3 |
| Mbandaka | 16 | 2.9 | 31.3 |
| Hadar | 16 | 10.3 | 93.3 |
| Kentucky | 15 | 7.1 | 46.7 |
| Montevideo | 9 | 0.0 | 0.0 |
| Thompson | 7 | 5.0 | 42.9 |
| Infantis | 6 | 0.0 | 0.0 |
| Ohio | 4 | 0.0 | 0.0 |
| Litchfield | 3 | 0.0 | 0.0 |
| Virchow | 1 | 5.9 | 100.0 |
| D | 4 | 0.0 | 0.0 |
| Enteriditis | 4 | 0.0 | 0.0 |
| E | 15 | 1.6 | 13.3 |
| Muenster | 10 | 0.0 | 0.0 |
| Anatum | 2 | 8.8 | 50.0 |
| Senftenberg | 2 | 2.9 | 50.0 |
| Orion | 1 | 0.0 | 0.0 |
| All isolates | 209 | 11.8 | 53.6 |
aPercentage of antimicrobial-resistance tests that were positive
Resistance among Salmonella isolates from bovine sources (n = 42) to ampicillin, ticarcillin, streptomycin, sulfamethoxazole, and tetracycline ranged between 60% and 69% (data not shown). The antimicrobials to which poultry isolates (n = 154) were most commonly resistant were tetracycline, streptomycin, and sulfamethoxazole; resistance ranged from 18% to 28%. Porcine isolates (n = 13) were mostly sensitive to all antimicrobials. The crude odds ratios in Table 4 show that Salmonella isolates from bovine sources were more commonly resistant to ampicillin, ticarcillin, kanamycin, streptomycin, sulfamethoxazole, tetracycline, and chloramphenicol than were those from poultry sources. The association between animal source and resistance to ampicillin, ticarcillin, streptomycin, sulfamethoxazole, and tetracycline, however, was only among those from veterinary diagnostic sources. Adjusted for submission source, odds ratios for resistance to ampicillin, ticarcillin, sulfamethoxazole, and chloramphenicol were significant (Table 4). When adjusted for animal source, submission source was a significant factor in resistance to ticarcillin, but was not significant with respect to resistance to any of the other antimicrobials (data not shown).
Table 4.
Antimicrobial resistance of bovine source Salmonella isolates compared with poultry source isolates, stratification by submission source
| Odds ratio (95% CI)
|
||||
|---|---|---|---|---|
| Stratified
|
||||
| Antimicrobiala | Crude | Monitoring | Diagnostic | Adjustedb |
| Ampicillin | 9.00 (4.20, 19.27) | 3.00 (0.67, 13.53) | 8.33 (2.65, 26.20) | 6.01 (2.49, 14.52) |
| Ticarcillin | 8.62 (4.04, 18.40) | 3.00 (0.67, 13.53) | 6.94 (2.27, 21.29) | 5.36 (2.23, 12.87) |
| Amoxicillin-clavulanic acid | 0.62 (0.17, 2.22) | 0 | 0.36 (0.08, 1.57) | — |
| Cephalothin | 0 | 0 | 0 | — |
| Ceftiofur | 0 | 0 | 0 | — |
| Gentamicin | 0 | 0 | 0 | — |
| Kanamycin | 9.41 (3.46, 25.63) | 2.43 (0.26, 22.58) | 0 | — |
| Streptomycin | 3.80 (1.87, 7.72) | 1.34 (0.31, 5.88) | 11.00 (3.09, 39.20) | — |
| Nalidixic acid | 0 | 0 | 0 | — |
| Sulfamethoxazole | 8.85 (4.12, 19.00) | 2.42 (0.54, 10.83) | 11.92 (3.59, 39.61) | 6.73 (2.81, 16.14) |
| Trimethoprim-sulfamethoxazole | 3.73 (0.23, 60.95) | 0 | 0 | — |
| Tetracycline | 5.76 (2.74, 12.11) | 1.44 (0.33, 6.35) | 11.92 (3.59, 39.61) | — |
| Chloramphenicol | 12.08 (4.87, 30.00) | 10.20 (2.02, 51.63) | 10.26 (2.09, 50.31) | 10.24 (3.04, 34.55) |
aResistance to ceftriaxone, amikacin, apramycin, and ciprofloxacin are not included because all isolates were susceptible
bMantel-Haenszel odds ratio (OR) adjusted for animal source is given when Breslow-Day test for homogeneity P > 0.05 and when non-zero OR is reported for both strata
Thirty-four isolates (16.3%) were resistant to 6 or more antimicrobials. All of these isolates belonged to serogroup B and almost all were of serovars Typhimurium or Typhimurium var. Copenhagen (91.2%). Twenty-two strains of phage type 104 were identified among the 54 S. Typhimurium and S. Typhimurium var. Copenhagen isolates. Five strains of phage type 104 were not resistant to any antimicrobials. These 5 isolates were from diverse origins and years, and the MICs of these isolates were between 2 and 8 dilutions below the breakpoint MICs.
The most common profile of resistance among the 209 Salmonella isolates was that which included the combined resistance to ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, tetracycline, and ticarcillin (ACSSuTTi) (Table 5). Isolates with this profile were largely of diagnostic origin (66.7% of 18) and this profile was found only among strains of serovars Typhimurium and Typhimurium var. Copenhagen. Of the 22 phage type 104 isolates, 13 (59.1%) had the ACSSuTTi resistance profile. Strains with the streptomycin and tetracycline (ST) resistance profile were mostly S. Hadar strains, while resistance to amoxicillin-clavulanic acid, ampicillin, cephalothin, and ticarcillin (AxACpTi) was found almost exclusively among isolates of S. Heidelberg from poultry sources (Table 5).
Table 5.
Number of Salmonella isolates with common antimicrobial resistance profiles and distribution of serovars (%) among isolates with resistance profiles
| Serovars
|
||||
|---|---|---|---|---|
| Resistance profile | n | Typhimurium and Typhimurium var. Copenhagen | Heidelberg | Hadar |
| ACSSuTTi | 18 | 100.0 | 0.0 | 0.0 |
| ST | 13 | 0.0 | 0.0 | 69.2 |
| T | 11 | 9.1 | 0.0 | 27.3 |
| AxACpTi | 11 | 0.0 | 90.9 | 0.0 |
| GSSu | 6 | 0.0 | 83.3 | 0.0 |
| GSu | 5 | 0.0 | 40.0 | 0.0 |
| AKSSuTTi | 5 | 100.0 | 0.0 | 0.0 |
Ax — amoxicillin-clavulanic acid; A — ampicillin; Cp — cephalothin; C — chloramphenicol; G — gentamicin; K — kanamycin; S — streptomycin; Su — sulfamethoxazole; T — tetracycline; Ti — ticarcillin
Discussion
Data analyses indicated that the proportions of Salmonella from food and food animals that are resistant to ampicillin, streptomycin, sulfamethoxazole, and tetracycline are high in Alberta, as they are in Canada (13). Percentage resistance captures the extent to which bacteria are resistant to all antimicrobials in the test panel. Poppe et al (13) found that of the 17 antimicrobial susceptibility tests conducted on 1336 Salmonella isolates from animals and foods of animal origin, 8.1% of the tests were negative for susceptibility. In that study, the list of antimicrobials used was not identical to that used in the present study, thus the 2 studies are not comparable. Many of the S. Hadar isolates examined in this study were resistant to antimicrobials, but the number of antimicrobials to which they were resistant was small. Consequently, although 93.3% of the S. Hadar isolates were resistant to 1 or more antimicrobials, the percentage resistance among S. Hadar was only 10.3%. In contrast, the percentage resistance for S. Typhimurium var. Copenhagen isolates was 3 times greater because, individually, they were resistant to more antimicrobials.
The results of this study suggest that within a sample of Salmonella isolates, the proportion of isolates that belong to serogroup B and certain serovars can influence the overall proportion of resistance within the sample. Farrington et al (14) suggested there may be a relationship between serogroup and antimicrobial resistance in Salmonella. In general, in this study, resistance was more common among isolates of serogroup B than among those of serogroups C and E, and more common among isolates of serogroup C than of serogroup E. The difference in resistance between isolates of serogroup B and other serogroups is more pronounced in this study than in the work of others (14,15). Salmonella Heidelberg from avian sources frequently possess large plasmids encoding antimicrobial resistance (16). Resistance to ampicillin, sulfamethoxazole, streptomycin, and tetracycline among S. Heidelberg isolated from poultry in the United States in 1987 and in Canada in 1993 was between 4% and 57%, respectively (16,17). These levels are similar to those observed among S. Heidelberg from poultry sources in the present study.
In this study, Salmonella from bovine sources were more commonly resistant to antimicrobials than were those from poultry, but this trend was only evident among isolates from veterinary diagnostic sources. Other researchers have reported that diagnostic isolates are more likely to be resistant to antimicrobials than are those originating from healthy animals at slaughter or from food products (13,18). We found that resistance did not, in general, differ significantly between isolates from veterinary diagnostic sources and those from monitoring sources when the animal source of isolates was also considered.
Profiles of antimicrobial resistance patterns were associated with specific serovars in this study: the ACSSuTTi profile was associated with serovars Typhimurium and Typhimurium var. Copenhagen, the AxACpTi profile was associated with S. Heidelberg, and the ST profile was associated with S. Hadar. Phage type 104 strains of S. Typhimurium commonly have an ACSSuT resistance profile, similar to the ACSSuTTi profile observed in phage type 104 strains of the present study (9,19–21). Strains of S. Typhimurium phage type 104 that are resistant to none of the antimicrobials used in testing are rarely reported in the literature. We observed a number of such strains, as did Poppe et al (22), who noted that among strains of S. Typhimurium phage type 104 there is a degree of diversity of resistance profiles, likely due to horizontal transfer of resistance genes.
Jones et al (23) advocated the value of MIC data, which provides detailed information about antimicrobial activity and are not prone to loss of relevance with changes in breakpoint guidelines over time. They argued that MIC data are crucial for surveillance and longitudinal research. In the present study, the maximum MIC for most antimicrobials among half of the Salmonella isolates was well below the MIC defined for resistance for those antimicrobials. In other words, the resistance to most antimicrobials would not have increased substantially if the breakpoint against which it was assessed was 1 or 2 MIC increments lower.
Estimates of the proportion of antimicrobial resistance reported in this study may not be a valid representation of the proportion among Salmonella from food animals and foods in Alberta due to the purposive isolate selection and small sample size. Furthermore, comparisons among clinical and active monitoring isolates, animal sources, serovars, and serogroups are suggestive but not conclusive. Nevertheless, antimicrobial sensitivity testing of this limited number of Salmonella isolates has provided some baseline information on the resistance of Salmonella to individual and multiple antimicrobials, and potential emerging trends in food animals and foods in Alberta. Future investigations of antimicrobial resistance in Salmonella from foods and food animals in the province should be based on a systematically and randomly sampled collection of isolates.
Acknowledgments
The authors thank Marg McFall and Louise Hawker from the Agri-Food Systems Branch of Alberta Agriculture, Food and Rural Development for assistance in susceptibility testing and data collection. CVJ
Footnotes
Financial support from Alberta Agriculture, Food and Rural Development is gratefully acknowledged.
References
- 1.Hein I, Schneck C, Knogler M, et al. Campylobacter jejuni isolated from poultry and humans in Styria, Austria: epidemiology and ciprofloxacin resistance. Epidemiol Infect. 2003;130:377–386. [PMC free article] [PubMed] [Google Scholar]
- 2.Aarestrup FM, Seyfarth AM, Emborg HD, Pedersen K, Hendriksen RS, Bager F. Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob Agents Chemother. 2001;45:2054–2059. doi: 10.1128/AAC.45.7.2054-2059.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith KE, Besser JM, Hedberg CW, et al. Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992–1998. N Engl J Med. 1999;340:1525–1532. doi: 10.1056/NEJM199905203402001. [DOI] [PubMed] [Google Scholar]
- 4.Winokur PL, Brueggemann A, DeSalvo DL, et al. Animal and human multidrug-resistant, cephalosporin-resistant Salmonella isolates expressing a plasmid-mediated CMY-2 AmpC beta-lactamase. Antimicrob Agents Chemother. 2000;44:2777–2783. doi: 10.1128/aac.44.10.2777-2783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aarestrup FM, Bager F, Jensen NE, Madsen M, Meyling A, Wegener HC. Resistance to antimicrobial agents used for animal therapy in pathogenic-, zoonotic-and indicator bacteria isolated from different food animals in Denmark: a baseline study for the Danish Integrated Antimicrobial Resistance Monitoring Programme (DANMAP) APMIS. 1998;106:745–770. doi: 10.1111/j.1699-0463.1998.tb00222.x. [DOI] [PubMed] [Google Scholar]
- 6.Marano NN, Rossiter S, Stamey K, et al. The National Antimicrobial Resistance Monitoring System (NARMS) for enteric bacteria, 1996–1999: surveillance for action. J Am Vet Med Assoc. 2000;217:1829–1830. [PubMed] [Google Scholar]
- 7.Wray C, Gnanou JC. Antibiotic resistance monitoring in bacteria of animal origin: analysis of national monitoring programmes. Int J Antimicrob Agents. 2000;14:291–294. doi: 10.1016/s0924-8579(00)00139-4. [DOI] [PubMed] [Google Scholar]
- 8.Franklin A, Acar J, Anthony F, et al. Antimicrobial resistance: harmonisation of national antimicrobial resistance monitoring and surveillance programmes in animals and in animal-derived food. Rev Sci Tech. 2001;20:859–870. doi: 10.20506/rst.20.3.1315. [DOI] [PubMed] [Google Scholar]
- 9.Sorensen O, Van Donkersgoed J, McFall M, Manninen K, Gensler G, Ollis G. Salmonella spp. shedding by Alberta beef cattle and the detection of Salmonella spp. in ground beef. J Food Prot. 2002;65:484–491. doi: 10.4315/0362-028x-65.3.484. [DOI] [PubMed] [Google Scholar]
- 10.National Committee for Laboratory Standards. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; Approved Standard. Wayne, Pennsylvania: National Committee for Clinical Laboratory Standards, 2002.
- 11.National Committee for Laboratory Standards. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; Approved Standard. Wayne, Pennsylvania: National Committee for Clinical Laboratory Standards, 1999.
- 12.National Antimicrobial Resistance Monitoring System. NARMS 1999 Annual Report. Last revised October 1, 2003. Available from http://www.cdc.gov/narms/annual/1999/html/narms_1999_annual.htm
- 13.Poppe C, Ayroud M, Ollis G, et al. Trends in antimicrobial resistance of Salmonella isolated from animals, foods of animal origin, and the environment of animal production in Canada, 1994–1997. Microb Drug Resist. 2001;7:197–212. doi: 10.1089/10766290152045084. [DOI] [PubMed] [Google Scholar]
- 14.Farrington LA, Harvey RB, Buckley SA, Droleskey RE, Nisbet DJ, Inskip PD. Prevalence of antimicrobial resistance in Salmonellae isolated from market-age swine. J Food Prot. 2001;64:1496–1502. doi: 10.4315/0362-028x-64.10.1496. [DOI] [PubMed] [Google Scholar]
- 15.D’Aoust J, Sewell AM, Daley E, Greco P. Antibiotic resistance of agricultural and foodborne Salmonella isolates in Canada: 1986–1989. J Food Prot. 1992;55:428–434. doi: 10.4315/0362-028X-55.6.428. [DOI] [PubMed] [Google Scholar]
- 16.Poppe C, McFadden KA, Demczuk WH. Drug resistance, plasmids, biotypes and susceptibility to bacteriophages of Salmonella isolated from poultry in Canada. Int J Food Microbiol. 1996;30:325–344. doi: 10.1016/0168-1605(96)00960-9. [DOI] [PubMed] [Google Scholar]
- 17.Lee LA, Threatt VL, Puhr ND, Levine P, Ferris K, Tauxe RV. Antimicrobial-resistant Salmonella spp isolated from healthy broiler chickens after slaughter. J Am Vet Med Assoc. 1993;202:752–755. [PubMed] [Google Scholar]
- 18.Altekruse SE, Elvinger F, Lee KY, et al. Antimicrobial susceptibilities of Escherichia coli strains from a turkey operation. J Am Vet Med Assoc. 2002;221:411–416. doi: 10.2460/javma.2002.221.411. [DOI] [PubMed] [Google Scholar]
- 19.Abouzeed YM, Hariharan H, Poppe C, Kibenge FS. Characterization of Salmonella isolates from beef cattle, broiler chickens and human sources on Prince Edward Island. Comp Immunol Microbiol Infect Dis. 2000;23:253–266. doi: 10.1016/s0147-9571(99)00079-x. [DOI] [PubMed] [Google Scholar]
- 20.Besser TE, Goldoft M, Pritchett LC, et al. Multiresistant Salmonella Typhimurium DT104 infections of humans and domestic animals in the Pacific Northwest of the United States. Epidemiol Infect. 2000;124:193–200. doi: 10.1017/s0950268899003283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szmolleny G, Kostyak A, Kovacs S, et al. Epidemiology and characterization of animal Salmonella enterica subspecies Enterica serotype typhimurium DT104 in Hungary. Acta Vet Hung. 2000;48:407–420. [PubMed] [Google Scholar]
- 22.Poppe C, Ziebell K, Martin L, Allen K. Diversity in antimicrobial resistance and other characteristics among Salmonella Typhimurium DT104 isolates. Microb Drug Resist. 2002;8:107–122. doi: 10.1089/107662902760190653. [DOI] [PubMed] [Google Scholar]
- 23.Jones RN. Detection of emerging resistance patterns within longitudinal surveillance systems: data sensitivity and microbial susceptibility. MYSTIC Advisory Board. Meropenem Yearly Susceptibility Test Information Collection. J Antimicrob Chemother. 2000;46:1–8. [PubMed] [Google Scholar]


