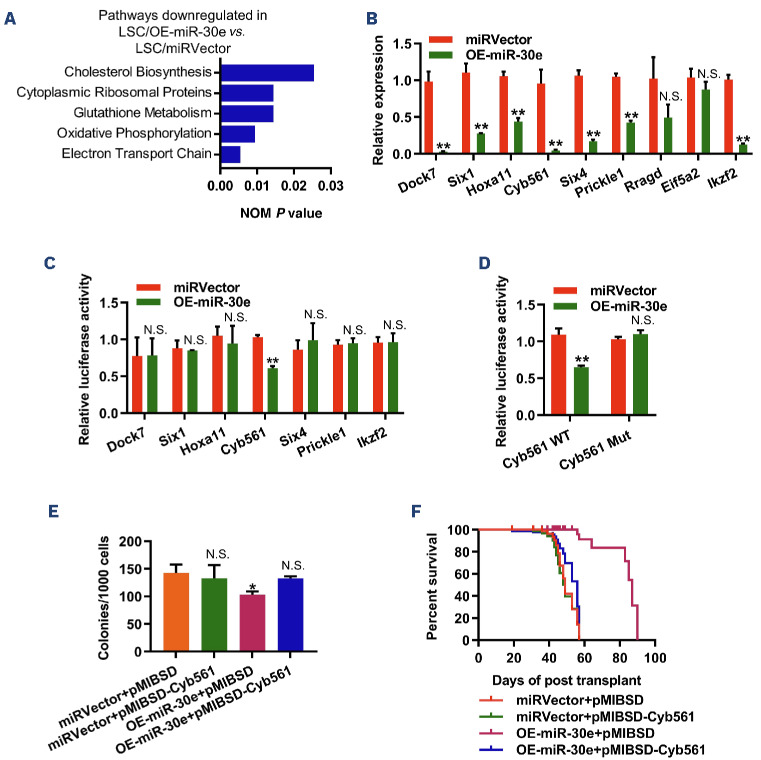
Figure 3.
RNA-sequencing analysis identifies potential targets of miR-30e-5p in L-GMP cells from KMT2A::MLLT3-driven leukemia. (A) Enrichment plot of downregulated gene sets in miR-30e-overexpressing leukemia stem cells (LSC), as determined by gene set enrichment analysis. RNA-sequencing data for 27,359 transcripts were used for the analysis. (B) Quantitative polymerase chain reaction analysis of candidate target genes in LSC-enriched granulocyte-monocyte progenitors (L-GMP) sorted from recipients transplanted with miRVector- or miR-30e-overexpressing acute myeloid leukemia cells. Results are normalized to Hprt expression and expressed relative to the expression of target genes in miRVector L-GMP (N=3). (C) Luciferase reporter assay to identify the true target of miR-30e-5p (N=3). (D) Cyb561 3’UTR luciferase reporter assay to identify the binding site of miR-30e-5p (N=3). (E) Overexpression of Cyb561 impaired the suppressive function of miR-30e on LSC (N=3). (F) Forced expression of Cyb561 impeded the prolonged survival of miR-30e on KMT2A::MLLT3-driven leukemia (N=7). Data are representative of two or three independent experiments. All data are represented as mean ± standard deviation. Two-tailed Student t tests were used to assess statistical significance (N.S.: not significant; *P<0.05; **P<0.01). miRVector: control; OE-miR-30e: cells with miR-30e overexpression; NOM: nominal; WT: wild-type; mut: mutated.