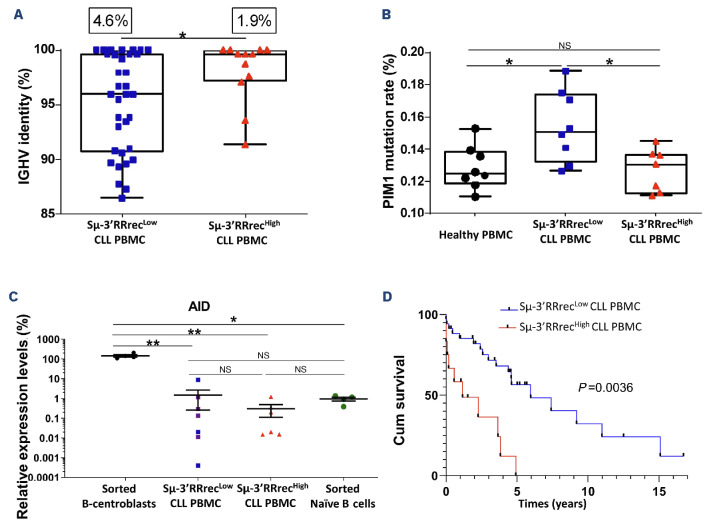
Figure 2.
Enrichment in unmutated chronic lymphocytic leukemia and poor prognosis of chronic lymphocytic leukemia patients with increased Sμ-3’RRrec counts. (A) Low Sμ-3’RR recombination (Sμ-3’RRrecLow) chronic lymphocytic leukemia (CLL) cases (N=34) had lower percentages of sequence identity with the reference sequence compared to the high homology of the variable region of the immunoglobulin heavy chain variable region (IGHV) segments in Sμ-3’RRrecHigh CLL (N=12). For each CLL group, the somatic hypermutation rate mean is indicated above the graph. (B) Sequence analysis of PIM1, activation induced-cytidine deaminase (AID) off-target gene. The mutation rate of PIM1 was significantly increased in Sμ-3’RRrecLow patients (N=8) compared to healthy peripheral blood mononuclear cells (PBMC) (N=8) and Sμ-3’RRrecHigh CLL (N=7). (C) AID transcripts, relative to CD19 transcripts, were lower in Sμ-3’RRrecLow CLL (N=7) and Sμ-3’RRrecHigh CLL (N=6) compared to normal B centroblasts (N=4) used as positive controls and comparable to AID transcript levels in sorted naïve B cells (N=4) used as negative controls. Purple dots correspond to mutated IGHV CLL samples. (D) Cumulative survival (Cum survival) time (years) without treatment (treatment-free survival [TFS]) for patients indicated shorter TFS in Sμ-3’RRrecHigh CLL (N=12) than Sμ-3’RRrecLow CLL (N=34). Graphs represent the mean ± standard error of the mean. Statistical analyses were performed using unpaired t test (A, B, C) or Χ2 test (D). 3’RR: 3’ regulatory region; NS: not significant; *P<0.05; **P<0.01; ***P<0.001.