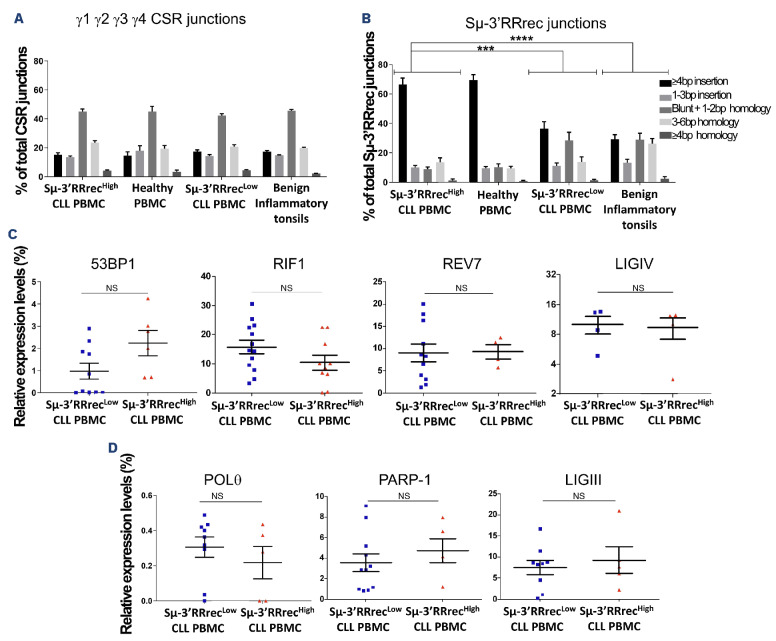
Figure 5.
Sμ-3’RRrec junction structural features are related to different lymphoid tissue imprints and discriminate between Sμ-3’RRrecLow and Sμ-3’RRrecHigh chronic lymphocytic leukemia. Structures at the Sμ-3’RR recombination (Sμ-3’RRrec) and class switch recombination (CSR) junctions were determined using CSReport by alignment to reference sequences for chronic lymphocytic leukemia (CLL) samples, healthy volunteer (HV) peripheral blood mononuclear cells (PBMC) and benign inflammatory tonsil cells. Structural features account for length in base pairs (bp) of nucleotide insertions at the joint, of short homology (microhomology) between acceptor and donor sequences and absence of insertions and homology (blunt) (A) Sμ-Sy1, SΜ-SY2, SΜ-SY3, SΜ-SY4 CSR joint structures were comparable between HV PBMC, benign inflammatory tonsils and CLL. (B) The Sμ-3’RRrec junction structure differed between CLL: Sμ-3’RRrecHigh samples were comparable to HV PBMC and predominantly exhibited junctions with long insertions (≥4 bp) while Sμ-3’RRrecLow CLL and benign inflammatory tonsils were enriched in Sμ-3’RRrec junctions with small microhomologies (1-2 bp) and blunt junctions. Quantification of transcripts coding for actors implicated in double strand break (DSB) repair by quantitative real-time polymerase chain reaction relative to CD19 transcripts (C) by non-homologous end joining (NHEJ) (53BP1, RIF1, Rev7 and LIGIV) and (D) by alternative end-joining (Alt-EJ) (PARP-1, POL9 and LIGIII). Statistical analyses were performed using the X2 test (A, B) or unpaired t test (C, D). 3’RR: 3’ regulatory region; NS: not significant; ***P<0.001; ****P<0.0001.