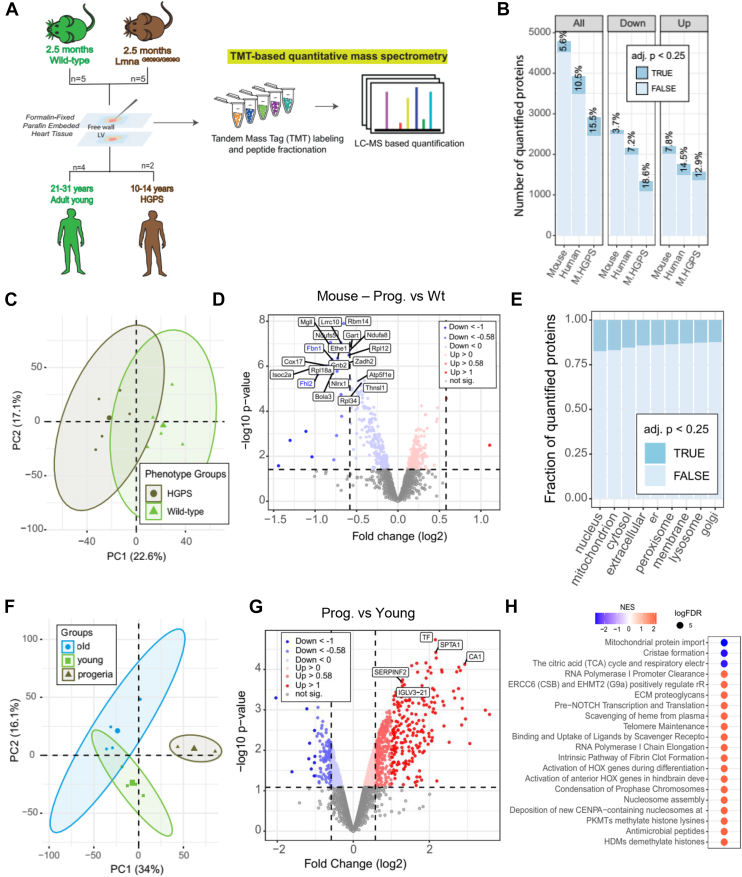
Fig. 4.
Impact of HGPS on cardiac tissue in mice and humans.A, schematic workflow of proteomic experiments using formalin-fixed paraffin-embedded (FFPE) left ventricles (free wall) of mice and humans. The proteome of LV tissue from age-matched (i) Wild-type and progeroid mice (LmnaG609G/G609G) (2.5 months old) and (ii) FFPE left ventricular tissue from young adults (25–31 years old) and two individuals diagnosed with HGPS (10 and 14 years old) was analysed by TMT-based quantitative mass spectrometry. Five male C57BL/6 mice were used for each group, Wild-type and progeroid (Lmna G609G/G609G). Four young male donors were used for the young group and one female and one male donor for the HGPS group. B, number of all and differentially (up and down-altered) quantified protein groups in each data set: mouse progeroid (M.HGPS) group (Wild-type vs. progeroid mice) and were added for comparison with previous data sets from mice and humans comparing young versus old. The percentage of significantly affected proteins (adj. p < 0.25) is indicated in dark blue. C, PCA plot showing the correlation of phenotype-related changes in the mouse proteome (mouse HGPS and Wild-type). D, volcano plot showing the significantly up (red) and down (blue) altered proteins (adj. p value <0.25 and absolute log2 fold change >0.58) comparing the progeroid phenotype with the Wild-type. Proteins not affected are shown in grey.E, compartmental analysis displaying the proportion of significantly altered proteins (adj. p < 0.25) by HGPS for each cellular compartment. In the plot, “er” means endoplasmic reticulum and “extracellular” represents ECM and ECM-associated proteins”. F, PCA plot showing the correlation among age-related alterations (old and young) and HGPS in human LV proteome. G, volcano plot displaying the dispersion of all quantified proteins according to their log2 fold changes and statistical significance (adj. p < 0.25) when comparing HGPS with young human LV tissue. H, gene set enrichment analysis (GSEA) display of the 20 most affected pathways during pathological aging in humans. GSEA was performed using WebGestalt (64). In the PCAs, the ellipses represent 95% confidence intervals. For each principal component (PC), the percentage of variance is indicated.