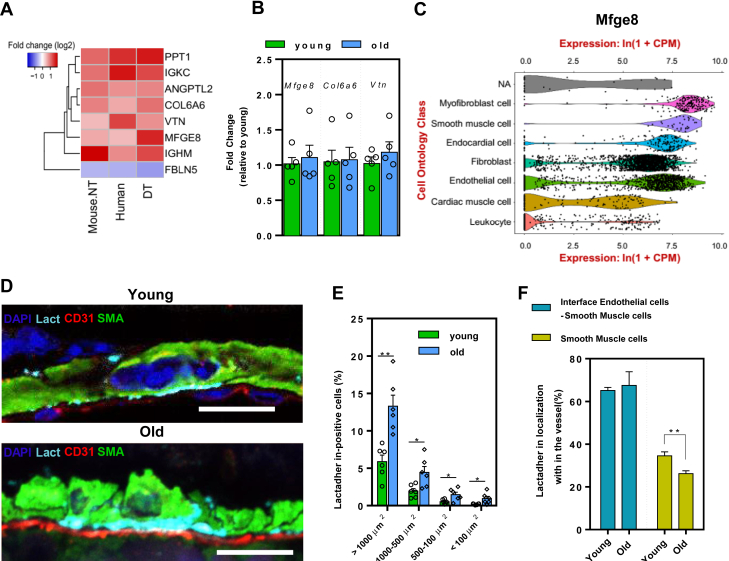
Fig. 5.
Validation of proteomic data and localization of lactadherin in LV tissue.A, heatmap showing the absolute log two fold change (FC) higher than 0.5 for the eight most statistically affected ECM proteins (p < 0.05 and absolute log2 FC > 0.5) and in three of four analysed proteomic datasets, in native mouse (NT), human NT and decellularized mouse LV tissues (DT). B, gene expression analysis by RT-qPCR for three of these ECM proteins selected from (a), in LV isolated from 3 months old (young) and 20 months old (old) C57BL/6 mice. The levels were normalized to the average expression level of the young mice. Results are Average ±SEM, n = 5, per group. C, Violin plot showing the expression levels of Mfge8 (lactadherin) in the heart based on single-cell data from Tabula Muris– FACS (30). D, representative images of immunofluorescence staining for lactadherin (Lact) (cyan), CD31 (red), smooth muscle actin (SMA) (green) and DAPI (blue) from LV sections of 3 months (young) and 22 months old (old) mice. Scale bar: 10 μm. E, bar plot shows the average of the percentage of lactadherin measured in the largest (>1000 μm2) to smallest (<100 μm2) blood vessels of young and old mouse LV tissue. Results are Average ± SEM, n = 3, per group. An average of 30 images for each section, two sections for each n were measured; ∗∗p < 0.01; ∗p < 0.05; Unpaired t test. F, bar plot showing localization of lactadherin in blood vessels. n = 3, per group (young and old). An average of 10 hit areas per section were analysed, two sections for each n. ∗∗p < 0.01; two-tailed un-paired student’s t test.