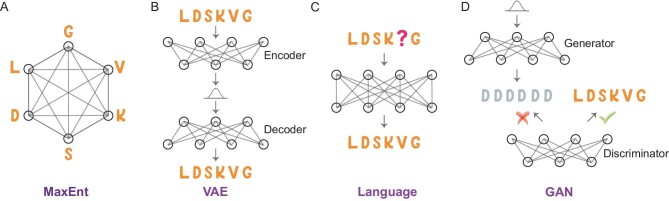
Figure 1.
Comparative illustration of generative models utilized for protein sequence modeling. (A) MaxEnt model: This model aims to delineate both the conservation of individual amino acids and their pairwise interactions, while concurrently making minimal assumptions by maximizing sequence information entropy. (B) VAE: A neural network that learns to encode data into a lower-dimensional latent space and then decode it back; after training, it can effectively generate new data that resemble the training set. (C) Language model: A masked language model employs a prediction-based mechanism that strives to accurately forecast the masked amino acid, thus learning the distribution of a corpus of protein sequences. (D) GAN: A framework utilizes two neural networks operating in tandem—a generator that creates new protein sequences, and a discriminator that evaluates them for authenticity.