Figure 4. Molecular modeling of alpha-1-antitrypsin (AAT)–glucocorticoid receptor (GR) complex.
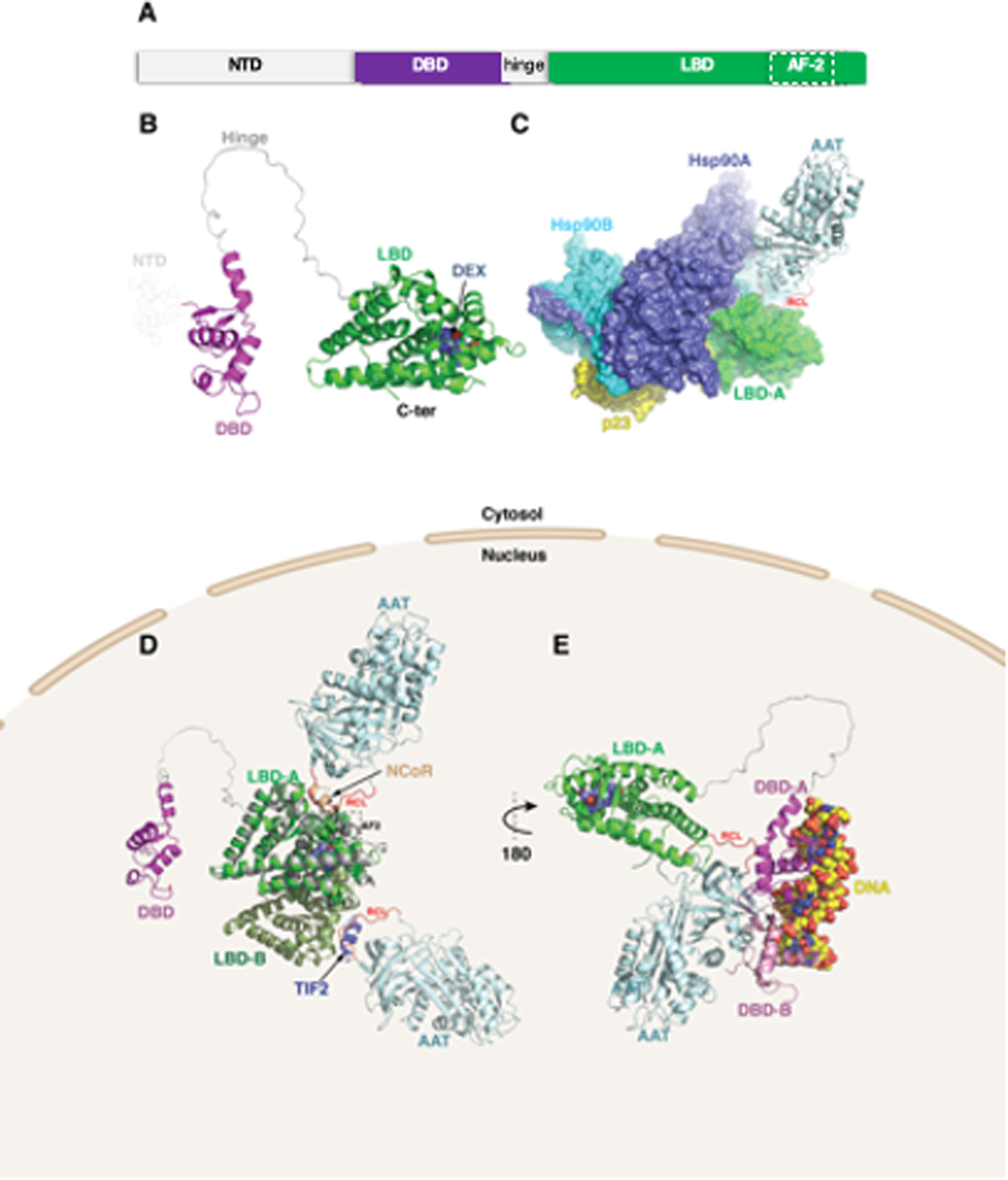
(A) GR is organized into three major domains: an intrinsically disordered N-terminal activation function-1 domain (NTD), a DNA binding domain (DBD), and a C-terminal ligand binding domain (LBD). (B) Structural modeling of the GR protein reveals the presence of the LBD (green) and DBD (magenta), linked by an unstructured hinge region (grey). The intrinsically disordered NTD was not modeled and is depicted by dotted lines. Dexamethasone bound to the LBD is shown as steel blue spheres. (C) AlphaFold2 docking simulations of LBD and AAT (PDB ID 3NE4) superimposed on the GR-Hsp90-p23 co-chaperone complex (PDB ID 7KRJ) show AAT interacting with the LBD via its RCL. (D) The same AAT–GR complex expanded to the LBD dimer (monomers labeled LBD-A and LBD-B). Superimposed are the nuclear coregulator proteins nuclear receptor corepressor (NCoR; derived from PDB ID 3H52) and transcriptional intermediary factor-2 (TIF2; derived from PDB ID 1M2Z) bound to the AF-2 site (broken rectangle) are shown as brown and slate-blue cartoons, respectively. (E) The second model using ClusPro docking shows the RCL of AAT interacting with the LBD of GR. In addition, the DBD is shown complexed with DNA (space-filling atoms), modeled using the GR DNA Binding Domain monomer – TSLP nGRE Complex (magenta; PDB ID 5HN5). Also shown (pink) is a second DBD formed by D-loop-mediated dimerization (PDB ID 5E69), having some degree of steric overlap with AAT. RCL (red)=reactive center loop.
