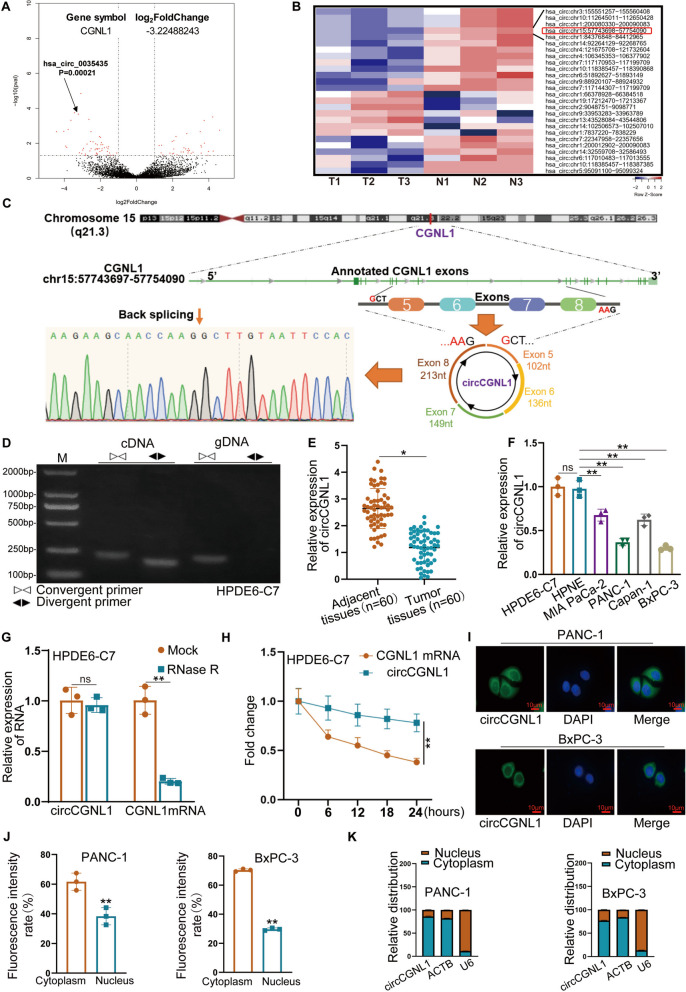
Fig. 1.
Identification and characteristics of circCGNL1 in PC cells and tissues. A, B Volcano plots [30] and a heatmap indicated that circCGNL1 (has_circ_0035435; chromosome 15, nt positions 57743698-57754090) was expressed at lower levels in PC tissues (log2FC = -3.22) than in matched normal tissues. C Schematic illustration of circCGNL1 formation via circularization of exons 5–8 of a CGNL1 mRNA transcript. The back splicing site was validated using Sanger sequencing. D Agarose gel electrophoresis was performed to detect the cyclization of circCGNL1 in HPDE6-C7 cells. E, F qRT-PCR assays were performed to detect circCGNL1 expression in PC tissues and different cell lines (MIA PaCa-2, PANC-1, Capan-1, BxPC-3, HPDE6-C7 and HPNE). G qRT-PCR analysis showing the expression levels of circCGNL1 and CGNL1 mRNA in HPDE6-C7 cells subjected to RNase R treatment. H RNA-expression levels of circCGNL1 and CGNL1 in HPDE6-C7 cells analyzed using qRT-PCR after treatment for 24 h with actinomycin D (2 μg/mL). I-K FISH and Subcellular-fractionation assays were used to determine subcellular localization of circCGNL1 in PANC-1 and BxPC-3 cells. Fluorescence intensity of nucleus and cytoplasm was measured via ImageJ. Scale bar = 10 μm. ACTB (cytoplasm) and U6 (nucleus) were served as internal controls. ns: no significance, *p < 0.05, **p < 0.01