Figure 2.
Dysregulation of immune cells in ME/CFS
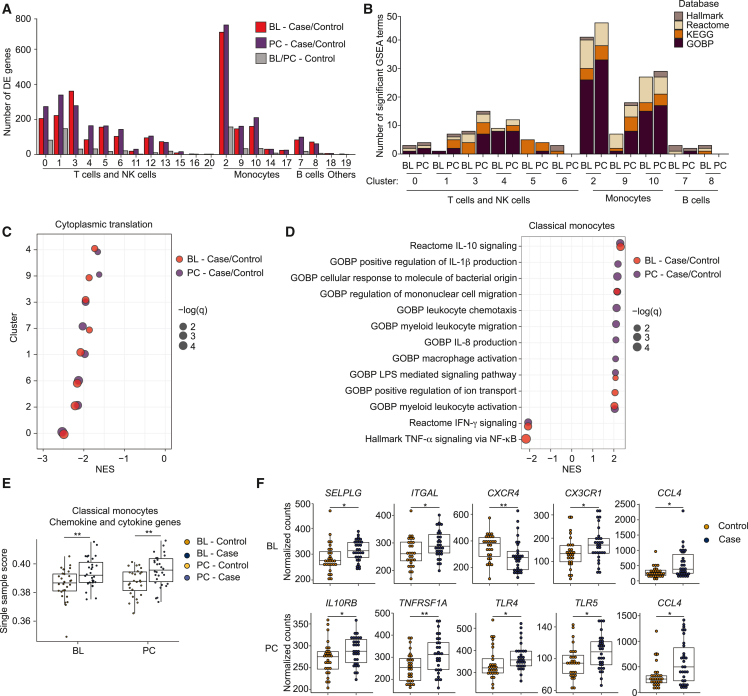
(A) Counts of differentially expressed (DE) genes (y axis) per immune cell cluster, comparing case and control cells at BL and PC, and compared between control cells at BL and PC.
(B) Counts of the strongest significantly enriched gene sets (excluding those related to translation and with an absolute normalized enrichment score [NES] <2) across the largest cell clusters.
(C) Representative GSEA gene set (GOBP cytoplasmic translation) showing lower detection of ribosomal proteins in major clusters (NES <0) at both BL and PC in ME/CFS (red and purple, respectively). Dots are sized to denote significance (q values); x axis indicates NES.
(D) GSEA results for classical monocytes (cluster 2) comparing patient and control cohorts at BL and PC (red and purple, respectively), focusing on gene sets related to chemokine/cytokine signaling.
(E) Single sample scores generated using GSEA of leading-edge genes from (D). ∗∗p < 0.01.
(F) Differential expression of genes associated with monocyte migration and differentiation at BL and PC in classical monocytes. ∗p < 0.05; ∗∗p < 0.01. Panels represent data from 28 healthy controls and 30 ME/CFS cases.