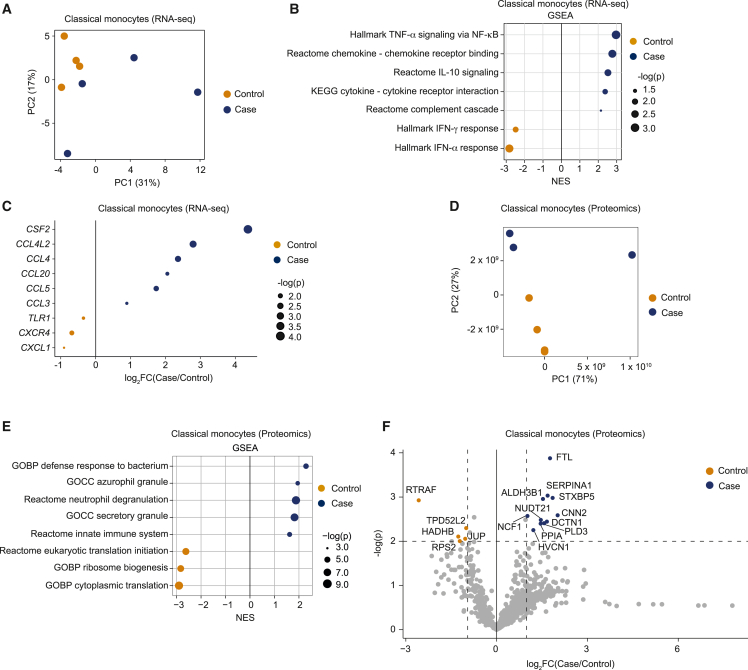
Figure 3.
Complete transcriptomics of classical monocytes
(A) PCA of 4 patient and 4 control transcriptomes from classical monocytes.
(B) Significantly enriched gene sets between cohorts by GSEA. Dots are sized to denote significance (adjusted p values); x axis indicates NES.
(C) Differentially expressed genes between cohorts. Dots are sized to denote significance (p values).
(D) PCA of 3 patient and 4 control proteomes from classical monocytes.
(E) Significantly enriched gene sets between cohorts by GSEA comparing proteome profiles of cases and controls; otherwise as in (B).
(F) Volcano plot of differentially expressed proteins between cohorts. Cohorts of 4 healthy controls and 4 ME/CFS cases (all females) at BL and PC were chosen for proteome (D–F) and transcriptome (A–C) profiling, respectively.