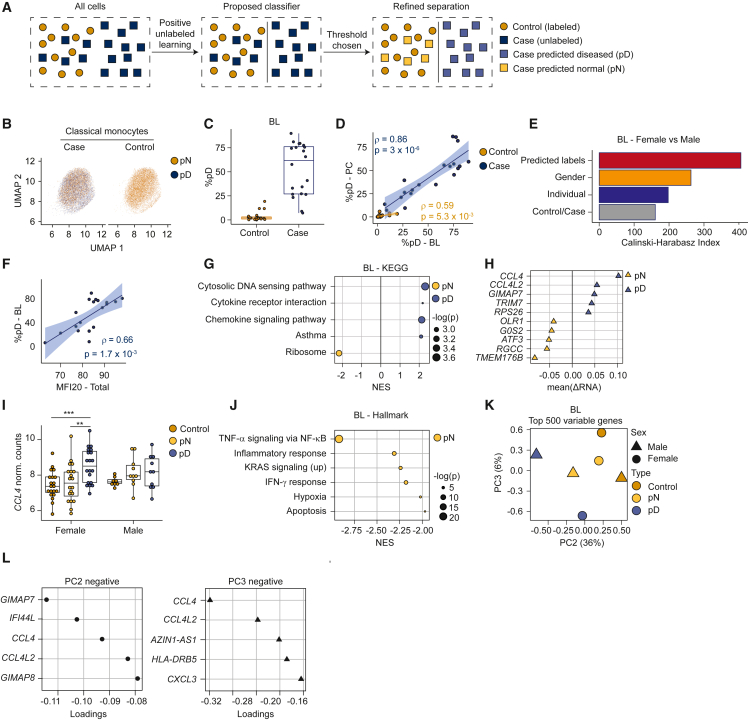
Figure 4.
Heterogeneity in classical monocyte cells from ME/CFS patients
Data represent BL female samples, unless described otherwise.
(A) Schema describing positive unlabeled learning strategy to stratify single-cell patient transcriptomes.
(B) UMAP for classical monocyte cells, tiled and colored by pD state.
(C) Percentage of pD cells per individual.
(D) Correlation (Spearman) between pD cells per individual, compared between BL and PC.
(E) CH index comparing clustering performance across different stratifications of the single-cell dataset (y axis).
(F) Correlation (Spearman) between MFI-20 score and percentage of pD cells.
(G) Gene sets most differentially enriched between pD and pN cells from cases. Dots are color-coded to indicate enrichment in pD (blue) or pN (yellow) cells; sizes indicate corrected p values.
(H) Genes (y axis) most differentially expressed (x axis) between pD and pN cells in an intrasample paired analysis.
(I) Expression of CCL4 (y axis) from individual samples, aggregating expression over pD and pN cells from cases and over all cells from controls (x axis and color-coded). ∗∗ p < 0.01, ∗∗∗ p < 0.001.
(J) Top gene sets differentially enriched between pD and pN cells, based on GSEA between each subset of cells paired by sample.
(K) Pseudobulk PCA from aggregated cells from control samples (yellow); pN and pD cells from case samples (green and blue, respectively), partitioned by sex.
(L) Genes contributing to negative values for PC2 (top) and PC3 (bottom) in (K).
(E, I, K, and L) Data from 28 healthy controls and 30 ME/CFS cases at BL, with partition based on sex in (I) and (K). (B–D) and (F–H) Data from the female cohort with 20 healthy controls and 20 ME/CFS cases at BL.