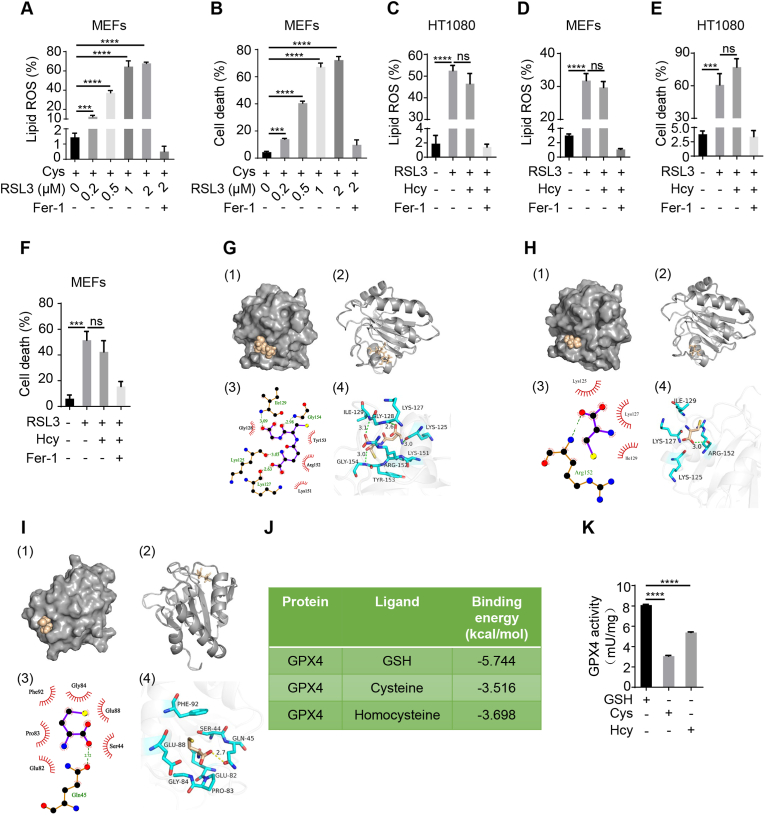
Fig. 4.
Cysteine and homocysteine inhibit ferroptosis by serving as the substrates for GPX4.
(A and B) Evaluation of lipid ROS and cell death for GPX4 inhibition in MEFs. Cells were treated with different concentrations of GPX4 inhibitor (RSL3, 2 μM) in the presence of cysteine (Cys) as indicated. After 4 h of treatment, lipid ROS (A) was assayed as described in Fig. 1 (B). Following 8 h incubation, cells were collected and subjected to evaluation of cell death (B) as described in Fig. 1 (C). (C and D) Assessment of lipid ROS for GPX4 inhibition or/and homocysteine (Hcy) treatment. HT1080 cells (C) and MEFs (D) were treated with RSL3 (2 μM) or in the presence of Hcy (400 μM) for 4 h, and subject to evaluation of lipid ROS as described in Fig. 1 (B). (E and F) Measurement of cell death in the context of GPX4 inhibition or combined with Hcy treatment. HT1080 cells (E) and MEFs (F) were treated as described in Fig. 4 (C and D) respectively for 8 h, cell death was determined as described in Fig. 1 (C). Treatment of ferroptosis inhibitor Fer-1 (10 μM) was the negative control. (G) Molecular docking analysis of binding pattern between GPX4 and GSH. Molecular surface of GPX4 in the region of GSH binding site in (1) and (2). Schematic drawing of the interaction between GSH and GPX4 in (3) and (4). Lys 125, Lys 127, Ile 129, and Gly 154 in the active cavity of GPX4 are key amino acid residues, which take responsibility for hydrophobic interaction between GSH and GPX4. (H) Molecular docking analysis of binding pattern between GPX4 and Cys. Molecular surface of GPX4 in the region of Cys binding site in (1) and (2). Schematic drawing of the interaction between Cys and GPX4 in (3) and (4). Arg 152 in the active cavity of GPX4 is key amino acid residue that is responsible for hydrophobic interactions between Cys and GPX4. (I) Molecular docking analysis of binding pattern between GPX4 and Hcy. Molecular surface of GPX4 in the region of Hcy binding site in (1) and (2). Schematic drawing of the interaction between Hcy and GPX4 in (3) and (4). Gln 45 in the active cavity of GPX4 is the key amino acid residue for the formation of hydrophobic interaction between Hcy and GPX4. (J) Binding energy analysis between GPX4 and its substrates. (K) Evaluation of GPX4 activity. Mouse liver extracts were prepared and subjected to assay of GPX4 activity in the presence of GSH (0.25 mM), Cys (0.25 mM), or Hcy (0.25 mM) as substrate respectively. Data represent as mean ± SD of three independent experiments, with significance determined by one-way ANOVA test. ***P < 0.001; ****P<0.0001; ns, nonsignificant.