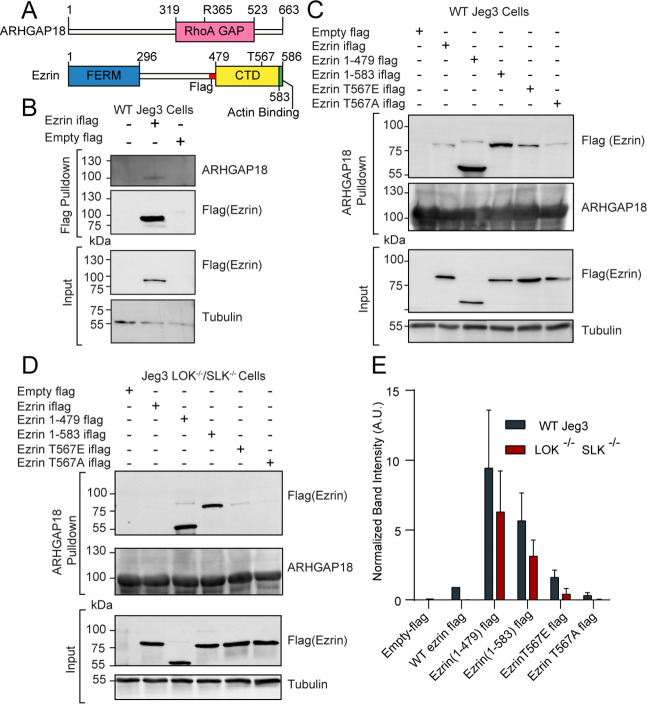
Figure 1. ARHGAP18 binds active, open ezrin through the FERM domain.
(A) Protein domain schematics of human ARHGAP18 and ezrin. (B) Western blot from pulldown experiment from passing WT-Jeg3 placental epithelial cell lysate over an anti-flag resin then blotting against flag-tagged ezrin and endogenous ARHGAP18. Ezrin tagged internally (iflag) within a flexible linker region at residue 479. (C) Western blot of ARHGAP18-bound resin pulldown experiment blotting against ezrin constructs expressed in WT-Jeg3 cells. (D) Western blot of ARHGAP18-bound resin pulldown experiment blotting against ezrin-constructs expressed in LOK-/-SLK-/--Jeg3 cells. (E) Quantification of normalized band intensity from all experiment presented in (C, D). Bars represents mean ± SEM; n = 4. Aggregated WT and LOK-/-SLK-/- conditions as populations were compared using a ratio paired t-test and found to be significantly different (p=0.0235).