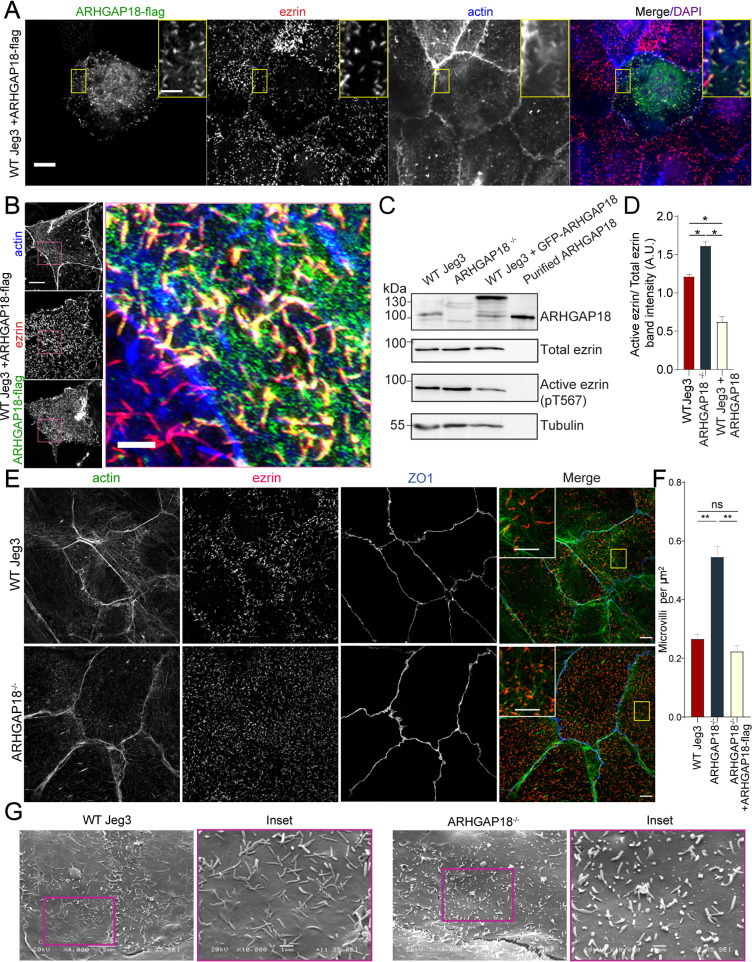
Figure 2. ARHGAP18 localizes to microvilli and alters ezrin T567 phosphorylation.
(A) Confocal immunofluorescence images of WT-Jeg3 cells expressing ARHGAP18-flag. Scale bar 10 μm; yellow box inset scale 2 μm. (B) Structured illumination microscopy (SIM) reconstructions of WT-Jeg3 cells expressing ARHGAP18-flag with zoom in on boxed area. Scale bar 10 μm; inset 2 μm. (C) Western blot showing CRISPR KO of endogenous ARHGAP18 and ezrin pT567 phosphorylation relative to total ezrin. Antibody against ARHGAP18 was confirmed in part by identification of endogenous ARHGAP18, GFP-ARHGAP18, and purified ARHGAP18 while showing no detection in KO cells. (D) Quantification of normalized band intensity from representative experiments presented in (C). Bars represent mean ± SEM; n = 4 paired t-test; *p≤0.05. (E) SIM images of WT vs. ARHGAP18-/- Jeg3 cells with ezrin as a marker for the abundance of microvilli at the apical surface. Scale bar 10 μm; yellow box inset scale 2 μm (F) Quantification of the density of microvilli on the surface of Jeg3 cells in WT, ARHGAP18-/-, and ARHGAP18 overexpression conditions. Microvilli counts per cell were normalized to the area of each cell to account for cell size variability. Bars represents mean ± SEM; n = 3 replicates; t-test; **p≤0.001. (G) Scanning electron microscopy (SEM) micrographs of WT-Jeg3 cells’ apical surface compared to those of ARHGAP18-/- cells. Left panels imaged at 4000×; scale 5 μm. Right panels imaged at 10,000× within of the boxed areas; scale 1 μm.