Abstract
Immunologically targeted therapies have revolutionized the treatment of inflammatory dermatoses, including atopic dermatitis and psoriasis. Although immunologic biomarkers hold great promise for personalized classification of skin disease and tailored therapy selection, there are no approved or widely used approaches for this in dermatology. This review summarizes the translational immunologic approaches to measuring treatment-relevant biomarkers in inflammatory skin conditions. Tape strip profiling, microneedle-based biomarker patches, molecular profiling from epidermal curettage, RNA in situ hybridization tissue staining, and single-cell RNA sequencing have been described. We discuss the advantages and limitations of each and open questions for the future of personalized medicine in inflammatory skin disease.
INTRODUCTION
Atopic dermatitis (AD) and psoriasis are two common chronic inflammatory dermatoses. Immune dysregulation drives AD and psoriasis pathogenesis (Di Cesare et al., 2009; Guttman-Yassky et al., 2011a, 2011b; Leung, 1999). Early studies in psoriasis showed an expansion of CD8+ T cells (Chang et al., 1994) capable of producing type 1 cytokines such as IFNγ and TNFα (Austin et al., 1999). IL-17 and IL-23 were subsequently found to be increased in psoriatic lesions and are now thought to represent a pathologic hallmark of this disease (Lee et al., 2004; Lowes et al., 2008; Piskin et al., 2006; Teunissen et al., 1998). These cytokines and other downstream proinflammatory signals mediate the characteristic changes of this disease (Chan et al., 2006; Liang et al., 2006; Teunissen et al., 1998).
AD lesional skin shows increased levels of IL-4 and IL-13, which are produced primarily by skin-infiltrating T helper (Th) 2 cells (Akdis et al., 1997; Renz et al., 1992; van der Heijden et al., 1991; van Reijsen et al., 1992). These type 2 cytokines are thought to be central to AD pathogenesis. Th22 cells may also be increased, resulting in excess IL-22 production (Gittler et al., 2012). IL-31, another type 2 cytokine, has also been implicated in AD pathogensis, particularly pruritus (Cheung et al., 2010; Dillon et al., 2004; Takaoka et al., 2006).
Despite the characteristic patterns of inflammation in most cases of AD and psoriasis, the Th1/Th17 versus Th2 paradigm may not apply neatly in all cases. There is evidence for intradisease heterogeneity within psoriasis and AD (Liu et al., 2022a; Tsoi et al., 2019; Wang et al., 2021a). Some studies have also suggested that molecular overlap may exist in some patients with psoriasis and AD (Moy et al., 2015). For example, some cases of AD may show an unexpected Th17/IL-23 signal (Brunner et al., 2018; Esaki et al., 2016; Koga et al., 2008), and this may be more common in pediatric individuals with AD and Asian individuals (Noda et al., 2015). Th1-driven inflammation may increase with chronicity in AD (Gittler et al., 2012). Th22 elevation has been reported in Black and African American patients with AD, with conflicting data on the role of Th1/17 inflammation in this population (Sanyal et al., 2019; Wongvibulsin et al., 2021). Overlapping Th2 and Th1/17 patterns have also been illustrated in palmoplantar pustulosis (McCluskey et al., 2022).
Biologic therapies have revolutionized treatment for psoriasis and AD. There are currently several cytokine-targeted biologic therapies approved for psoriasis, including TNFα inhibitors, IL-23 (p19) inhibitors, an IL-12/IL-23 (p40) inhibitor, IL-17A inhibitors, an IL-17 receptor (IL-17RA) inhibitor, and an IL-36 receptor (IL-36R) inhibitor. An oral TYK2 inhibitor was also recently approved for psoriasis. There are two currently approved biologics for AD in the United States: dupilumab, an IL-4Ra inhibitor (blocking the activity of both IL-13 and IL-4), and tralokinumab, an IL-13–specific inhibitor. In addition, two oral Jak inhibitors, upadacitinib and abrocitinib, which inhibit signaling downstream of many cytokines, were recently approved for AD. There are also numerous clinical trials for novel AD biologics, including drugs targeting IL-22, IL-31, and TSLP, among others.
Biologic therapies blocking the IL-17/IL-23 axis have shown 75% improvement in PASI-75 in approximately 75% of patients (Armstrong et al., 2020). In AD, IL-4Ra blockade with dupilumab results in a 75% improvement in the Eczema Area and Severity Index (EASI) 75 in approximately 50% of patients (Silverberg et al., 2021). Presently, medication choice is largely based on population-level efficacy data, medical comorbidities, and physician preference but, in most cases, is ultimately relies up trial and error (Aggarwal et al., 2022). Given the growing number of AD and psoriasis biologics aimed at different molecular targets, there is a need for approaches to assist with rational biologic treatment selection.
Biomarkers can be used for many purposes in inflammatory skin disease: to aid in diagnosis, to define clinical subtypes, to predict disease progression or severity, to monitor treatment response, or to inform therapy selection. Biomarkers for predicting disease progression or severity and monitoring therapy response in inflammatory skin disease have been reviewed elsewhere (Corbett et al., 2022; Ramessur et al., 2022; Renert-Yuval et al., 2021). In this paper, we focus on immune biomarkers that aim to inform biologic therapy selection and, in some cases, aid in the diagnosis of clinically/histopathologically indeterminate rashes (CIRs) (Figure 1). The notion of distinct clinical endotypes, which has been proposed in AD (Czarnowicki, 2019), correlates demographic factors such as age and ethnicity with underlying immunology and could be used to inform therapy selection. However, in this study, we focus on methods for selection of the best therapeutic target for individuals based on their unique immunologic profiles. In addition, although genetic polymorphisms; microbiome-based, metabolomic, or lipidomic biomarkers; as well as imaging-based biomarkers have been explored in inflammatory skin disease, we have chosen to focus on RNA- and protein-based immunologic biomarkers, which have the richest body of literature and the most direct relevance to therapy selection in inflammatory skin disease.
Figure 1. RNA- and protein-based biomarkers for biologic therapy selection and diagnosis.
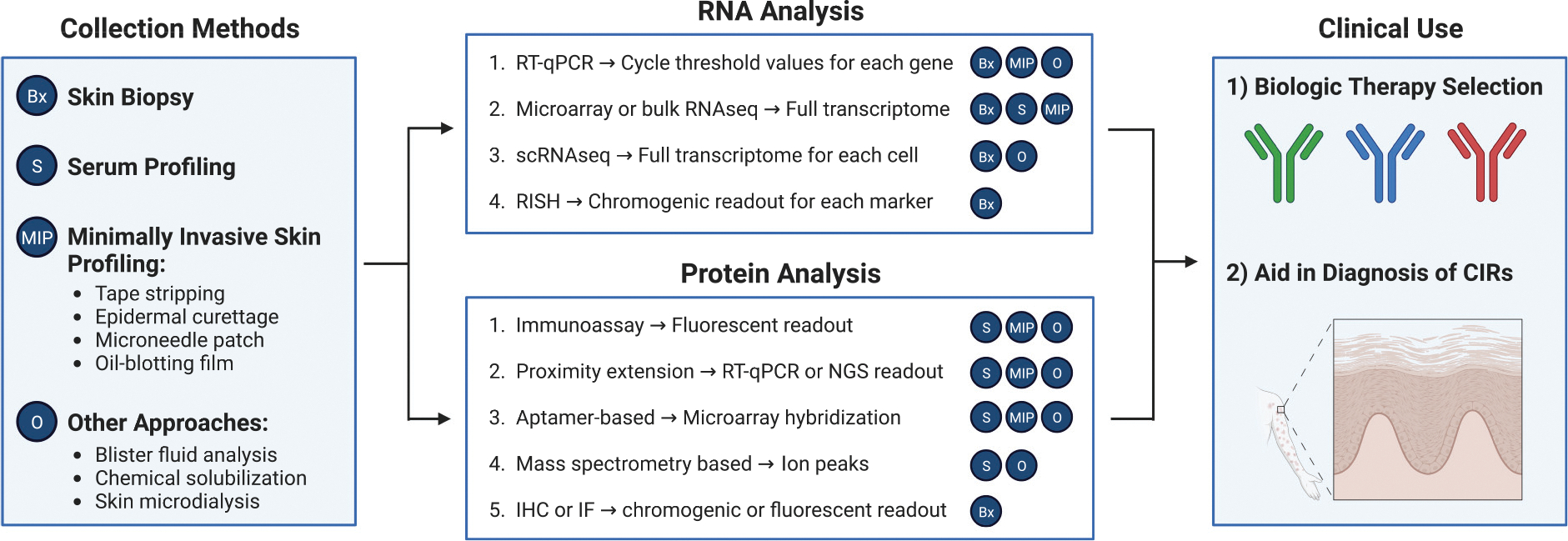
Various methods exist for collecting biomarkers from the skin, including shave or punch biopsy or minimally invasive approaches such as tape stripping, superficial epidermal curettage, or microneedle patches. In addition, systemic biomarkers from the blood can be measured. RNA analysis techniques include RT-PCR, microarray, sequencing approaches, or in situ analysis using hybridization probes. Proteins can be analyzed by IHC/IF, immunoassay, MS-based methods, proximity extension, or aptamer-based methods for high-throughput proteomics. Each method of analysis is labeled with compatible collection methods on the basis of references in this review. The RNA or protein levels of different immunologic molecules or other molecular correlates could be used for rational selection of biologic therapy and to aid in the diagnosis of CIRs. CIR, clinically/histopathologically indeterminate rash; IF, immunofluorescence; IHC, immunohistochemistry; RISH, RNA in situ hybridization; scRNA-seq, single-cell RNA sequencing; MS, mass spectrometry.
SKIN BIOMARKERS
Early studies used microarray profiling of skin biopsies to characterize transcripts enriched in psoriasis (Oestreicher et al., 2001; Suárez-Fariñas et al., 2012; Zhou et al., 2003), AD (Gittler et al., 2012; Guttman-Yassky et al., 2009; Nomura et al., 2003; Rodríguez et al., 2014), and shared gene signatures between psoriasis and AD (Choy et al., 2012). Other studies used RNA sequencing to identify differentially expressed genes in AD (Suárez-Fariñas et al., 2015) and psoriasis (Gudjonsson et al., 2010; Li et al., 2014; Swindell et al., 2013; Tsoi et al., 2015) and define subtypes of psoriasis on the basis of gene expression signatures (Ainali et al., 2012; Swindell et al., 2012). Several studies proposed classifiers to aid in the diagnosis of AD and psoriasis (Guttman-Yassky et al., 2009; Inkeles et al., 2015; Quaranta et al., 2014), including validation with prospective samples (Garzorz-Stark et al., 2016) and classification based on gene signatures from uninvolved skin (Tsoi et al., 2019).
Gene expression profiling of skin biopsies has also been employed to monitor response to treatment in AD (Beck et al., 2014; Guttman-Yassky et al., 2019a; Möbus et al., 2021) and psoriasis (Johnston et al., 2014; Krueger et al., 2019; Sofen et al., 2014). In one small-scale study in psoriasis, baseline levels of IL20, IL21, and p40 mRNA correlated with response to IL-12/23 inhibition (Gedebjerg et al., 2013). However, no large-scale studies have shown the ability of bulk transcriptomic profiling of the skin at baseline to predict response to biologic therapy. In AD, baseline Th2 biomarkers in the skin did not correlate with improvement in EASI or pruritis scores after treatment with IL-4Rα blockade (Beck et al., 2014). In a combined analysis of gene expression data of psoriatic lesions from many studies, authors achieved robust prediction of PASI improvements using gene expression values from 2 to 4 weeks of treatment but poor predictive accuracy using baseline measurements (Rosa da et al., 2017).
Proteomic analysis of skin biopsies has identified biomarkers in psoriasis (Carlén et al., 2005) and AD (Noh et al., 2016; Pavel et al., 2020), including immunohistochemistry (IHC)- and immunofluorescence (IF)-based methods, to classify AD and psoriasis skin with reasonable accuracy (D’Erme et al., 2015; Garzorz-Stark et al., 2016). To our knowledge, no studies have evaluated the ability of proteomic profiling of the skin to predict response to biologic therapy.
BLOOD BIOMARKERS
Many studies have uncovered alterations in circulating immune cells and levels of serum proteins in psoriasis and AD. Serum biomarkers have been useful to predict disease severity and progression and to monitor response to treatment, including in clinical trials (Beck et al., 2014; Guttman-Yassky et al., 2019a; Kim et al., 2018). The use of serum biomarkers for these purposes has been reviewed earlier (Mikhaylov et al., 2021a; Pourani et al., 2022; Sobolev et al., 2022).
Circulating biomarkers could aid in the selection of biologic treatment or the diagnosis of CIRs. High-throughput techniques such as proximity extension assays and aptamer-based methods have been employed to measure serum protein levels in AD and psoriasis (Brunner et al., 2017; Wang et al., 2017), in addition to more traditional immunoassays (Kolbinger et al., 2017). In one study, authors found that enhanced NF-κB signaling in immune cells from the blood of patients with psoriasis before therapy initiation was associated with poor response to TNFα inhibition (Andres-Ejarque et al., 2021). In an analysis of serum biomarkers from Chinese patients with AD before and after dupilumab treatment, one group found that baseline levels of three proteins–CD25/sIL-2Rα, IL-31, and IL-36β–had a modest correlation with response to treatment (Wu et al., 2023). Serum biomarkers have been used to classify adult and pediatric patients with AD into distinct clusters on the basis of their biomarker profiles (Bakker et al., 2022; Thijs et al., 2017), but it remains to be determined whether this classification could be used to predict response to biologic therapy. One recent study showed that dupilumab was equally effective in extrinsic-type (elevated IgE) AD and intrinsic-type (normal IgE) AD (Gelato et al., 2023).
MINIMALLY INVASIVE AND OTHER INNOVATIVE APPROACHES TO MEASURE IMMUNOLOGIC BIOMARKERS IN THE SKIN FOR BIOLOGIC THERAPY SELECTION
Tape strip profiling
Tape stripping is a minimally invasive method to profile the superficial epidermis. Adhesive tape strips are repeatedly applied to a patient’s skin, each time sampling a deeper portion of the stratum corneum (Figure 2). Approximately 20 tape strips are thought to be necessary to reach the granular cell layer of the epidermis (Kim et al., 2019), as each tape strip removes approximately one layer of corneocytes. Most studies to date have used less than 20 tape strips (Hughes et al., 2021). The procedure elicits only mild discomfort and/or erythema.
Figure 2. Skin sampling and processing for different investigational techniques for personalized therapy selection in inflammatory skin disease.
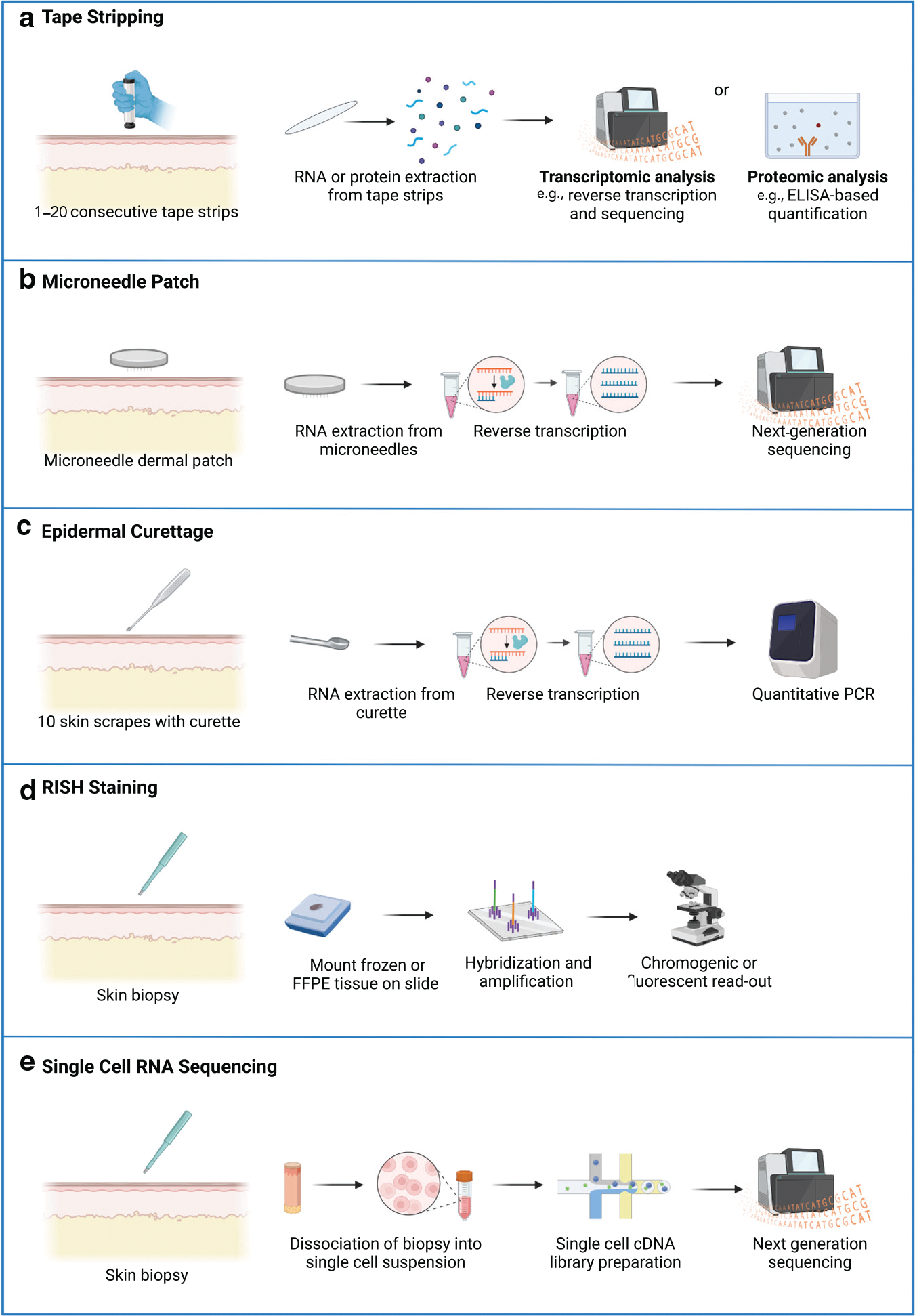
(a) For tape stripping, an applicator is applied to the skin for consecutive tape strips. RNA or proteins are extracted from the tape strips and quantified. (b) For the microneedle-based dermal biomarker patch (Mindera Health, San Diego, CA), a microneedle patch which has probes extending into the superficial dermis is applied to the skin. RNA is extracted, reverse transcribed, and sequenced. (c) For molecular profiling from epidermal curettage (Castle Biosciences, Friendswood, TX), the skin is scraped with a curette, and RNA is isolated from the skin sample and analyzed by RT-PCR. (d) For RISH, a skin biopsy is obtained. A probe complementary to the RNA molecule of interest is added to the tissue slide, and a chromogenic or fluorescent marker is added in the detection step. (e) For single-cell RNA sequencing, a biopsy is obtained, and the skin sample is dissociated into single cells. Single-cell cDNA library preparation is performed, and the cDNA library is sequenced using next-generation sequencing. FFPE, formalin-fixed, paraffin-embedded; RISH, RNA in situ hybridization.
Early studies of tape stripping used various techniques to quantify the mRNA levels of immune-related genes (Benson et al., 2006; Morhenn et al., 1999; Wong et al., 2004). He et al. (2021) performed RNA sequencing on tape strips from patients with AD, patients with psoriasis, and healthy controls. The authors showed that the skin of patients with AD had increased Th2-related transcripts (including IL13, CCL17, and CCL18) and that psoriasis skin had elevated Th17-related (IL17A/F) and innate (IL36A/IL36G) transcripts. They also showed that high levels of nitric oxide synthase 2 (NOS2) transcripts alone could identify psoriasis samples. Transcriptomic profiling of tape strip samples has also been applied to study pediatric AD (Guttman-Yassky et al., 2019b; Pavel et al., 2021) and hand eczema (Sølberg et al., 2022).
Dyjack et al. (2018) used tape stripping to identify heterogeneity within patients with AD, defining a group of type-2-high patients with elevated expression of IL13, IL4R, CCL22, and CCR4 and more severe eczema. Mikhaylov et al. (2021b) studied transcript levels from AD tape strips before and after dupilumab treatment, noting decreases in chemokine genes, including CCL13, CCL17, and CCL18, and increases in barrier-related transcripts after treatment toward the levels of healthy controls. Some gene expression changes correlated with responses to dupilumab treatment for all patients.
Investigators have also studied tape strip samples using proteomic approaches. Inoue et al. (2011) performed ELISA on tape strip samples to show that IL-18 levels are higher in the skin of patients with AD than in the skin of healthy controls. Méhul et al. (2017) used mass spectrometry and multiplex ELISA to define a proteomic signature of psoriatic lesions, and the same group also showed that proteomic profiling of tape strips could differentiate between psoriasis and cutaneous T-cell lymphoma (Méhul et al., 2019). In another study, authors found that the levels of IL-36γ quantified by ELISA from tape strip samples could accurately differentiate between psoriasis and AD (elevated in psoriasis), even in clinically challenging cases (Berekméri et al., 2018). Multiplex immunoassays have been used to quantify a larger number of protein biomarkers from tape strip samples of adult and pediatric patients with AD (Clausen et al., 2020; Hulshof et al., 2019; McAleer et al., 2019), including in response to dupilumab (He et al., 2020a). Mass spectrometry–based analysis of tape strip samples has also been used to compare protein biomarkers in the skin of patients with AD with and without food allergy (Goleva et al., 2020; Leung et al., 2019).
Tape stripping has several advantages for potential applications in personalized molecular profiling. The first is that it is minimally invasive, allowing for a higher number of sites or frequency of sampling and less discomfort for the patient. Second, direct comparisons between tape stripping and biopsy samples have shown that tape stripping may enrich for expression changes within the epidermis (Dyjack et al., 2018; Tsoi et al., 2022).
Limitations of tape stripping include that it predominantly samples the stratum corneum (and possibly the upper granular cell layer). One study found that proteomic profiling of tape strips was less sensitive to detecting cytokine changes in AD skin compared with RT-PCR of biopsy samples (Simonsen et al., 2021). In addition, there are reported difficulties in standardizing tape stripping on the basis of sampling procedure (number of tape strips and amount of pressure applied) and characteristics of the sampled skin, such as stratum corneum thickness and skin hydration (Bashir et al., 2001; Berekméri et al., 2019). Processing samples for RNA sequencing or proteomics requires specialized equipment, specific expertise, and data normalization, making it potentially expensive and limiting clinically relevant turnaround times (Table 1).
Table 1.
Comparison of Investigational Approaches for Personalized Therapy Selection in Inflammatory Skin Disease
| Tape Stripping | Dermal Biomarker Patch (Mindera Health) | Epidermal Curettage (Castle) | RISH Staining | scRNA-seq | |
|---|---|---|---|---|---|
| Sampling technique | 1–20 tape strips applied to the skin | Dermal biomarker patch applied to the skin for 5 minutes | 10 scrapes with curette | Skin biopsy | Skin biopsy |
| Layers of skin samples | Stratum corneum | Epidermis and upper dermis | Not publicly disclosed | Epidermis and dermis | Epidermis and dermis |
| Cryopreservation necessary | Yes. Must store tape strips at –80 °C | No. Can be stored at 4 °C and processed within 72 hours | Not publicly disclosed | No | Samples typically processed fresh |
| Likelihood of scar | Low | Not publicly disclosed | Not publicly disclosed | High | High |
| Integration into clinical workflow1 | Can be collected relatively rapidly; requires specific expertise, normalization, and possibly slow turnaround time. | Can be collected relatively rapidly. Turnaround time and cost of commercial service not yet clear. | Can be collected relatively rapidly. Turnaround time and cost of commercial service not yet clear. | Integrates well with IHC workflow in dermatopathology laboratories with rapid turnaround for specific targets. | Resource-intensive sample processing, sequencing, and data analysis. |
| Throughput/number of markers | Full transcriptome (for RNA-based methods) | Full transcriptome | 28 transcripts | One marker per staining; multiple markers can be batched | Full transcriptome |
| Requires sequencing | Yes | Yes | No | No | Yes |
| Data analysis | Computational pipelines for microarray or RNA- sequencing data. | Computational pipelines with proprietary algorithms. | Comparison of count threshold values from qPCR. | Manual or automated quantification of positive cells on microscopy slide. | Computational pipelines for scRNA-seq data. |
| Histologic analysis inherent | No | No | No | Yes | No |
Abbreviations: RISH, RNA in situ hybridization; scRNA-seq, single-cell RNA sequencing.
Cost will also be an important factor in comparing the feasibility of workflow integration, but there are not enough data at this time to compare costs between methods.
Microneedle-based dermal biomarker patch
Microneedles are micron-sized needles that can be used to sample RNA or protein from the skin (Ibrahim et al., 2022; Kim et al., 2022) or interstitial fluid (Samant et al., 2020; Wang et al., 2021b). One approach that is being commercially pursued (Mindera Health) to collect material for biomarker analysis from the skin uses microneedle technology through a dermal biomarker patch (DBP) (Ibrahim et al., 2022). Their DBP is lined with 100 square pyramidal microneedles with a 200 × 200 μm base and 750 μm depth. Microneedles are treated with DNA oligonucleotides to enhance the capturing of mRNA. The DBP is applied to the skin and left on for 5 minutes. The microneedles sample into the upper dermis to a depth of 350–400 μm. Subsequent RNA sequencing showed gene detection comparable with that of the RNA sequencing of punch biopsy specimens.
This technology was used in combination with a machine-learning algorithm to assess biologic responses in patients with psoriasis (Bagel et al., 2021; Mindera Health). DBPs were applied at baseline, and the change in PASI was assessed after 12 weeks of treatment with either an IL-23 inhibitor, an IL-17 inhibitor, a TNFα inhibitor, or an IL-12/23 inhibitor. The authors trained a classifier to predict response to IL-23 inhibitors in a subset of the patients, and they trained IL-17 and TNFα inhibitor response classifiers on the basis of previously generated gene expression data. Their classifiers achieved high positive predictive value (PPV) for response to therapy, with PPV values of 93.1, 92.3, and 85.7% for IL-23, IL-17, and TNFα inhibitor classifiers, respectively, when applied to high-PASI (PASI > 8) patients.
DBP appears less invasive than a traditional skin biopsy. Compared with tape stripping, microneedles can sample down into the dermis. One group showed a higher concentration of RNA in samples collected from microneedle patches than from tape stripping (Kim et al., 2022).
There are limitations to microneedle-based approaches. Applying microneedles to inflamed skin could lead to irritation and/or discomfort. Also similar to tape stripping, transcriptional profiling from microneedles requires specialized equipment and expertise to collect and analyze the data, which may restrict its use to a centralized or commercial setting. The cost and turnaround time of this process are not fully clear, but both could be potential limitations. Whether insurance will cover such approaches, including the Mindera system, is also not yet clear. Finally, because the epidermis is breached, there is potential for scarring.
Molecular profiling from epidermal curettage samples
Another minimally invasive way to collect skin samples is by curetting superficial epidermal tissue. One approach that is being pursued commercially (Castle Biosciences, Friendswood, TX) uses this sample collection technique for molecular profiling of 28 psoriasis and AD-related genes using RT-PCR (Quick et al., 2022). This approach was able to detect gene expression differences between AD and psoriasis.
Molecular profiling from epidermal curettage has the advantage of being minimally invasive, although it is possible that the depth of sampling might vary on the basis of the end user and anatomic location. If the dermo–epidermal junction is not breached, this technique would be expected to be nonscarring. In this case, the relative ease of sample collection would also allow for more frequent sampling or sampling at more sites.
Limitations include the specialized technology and expertise required for sample processing and data analysis, which also necessitate sample processing and analysis at a centralized location. In addition, no data are reported on the reproducibility of the collection method between operators or sites. The breadth of potential biomarkers is also limited by the current approach. Turnaround time, cost, and payor considerations are not yet fully clear for the Castle Bioscience approach under development.
RNA in situ hybridization staining
Biomarkers can be measured directly in skin biopsies using techniques such as IF and IHC. Although promising in concept, IHC and IF for secreted proteins such as cytokines have proven difficult because of often high background staining (Chen et al., 2023; Cohen et al., 2020; Miranda et al., 2021; Moy et al., 2015). RNA in situ hybridization (RISH) detects specific mRNA molecules by hybridization of complementary 18–25 bp probes to tissue sections, amplification of the signal, and detection using a chromogenic or fluorescent molecule (Wang et al., 2012). RISH can be performed on skin biopsies, including frozen and formalin-fixed, paraffin-embedded diagnostic specimens. The location of the desired mRNA can then be visualized in the histologic section. RNAScope is a commonly used, commercially available system for performing RISH. With RNAScope, signal detection and amplification require tandem binding of distinct probes, and thus the assay is considered to be highly specific.
RISH has been used to detect disease-relevant cytokines in AD and psoriasis biopsies. Our group used chromogen-based RISH (with the RNAScope kit, Advanced Cell Diagnostics, Hayward, CA) in a retrospective study to determine the levels and degrees of heterogeneity of druggable cytokines in AD and psoriasis biopsies (Wang et al., 2021a). The study focused on established treatment targets, including IL4, IL12B (IL-12/23 p40), IL13, IL17A, IL17F, IL23A (IL-23 p19), and TNF (TNFα); emerging AD therapeutic targets including IL22 and IL31; and established psoriasis biomarkers NOS2 and IFNG. We found that staining was able to discriminate between psoriasis (generally NOS2+ IL17A+), AD (generally NOS2− IL13+), and healthy control samples (negligible staining).
IL17A and IL13 were most commonly detected at significant levels in psoriasis and AD, respectively. IL12B, IL23A, and IL17F were also increased in many cases of psoriasis. Expression of IL4 was also present in AD but was generally present at markedly lower levels than that of IL13, consistent with earlier studies (Tsoi et al., 2019). Within each disease, we also found distinct molecular heterogeneity, and the predominantly expressed druggable cytokines varied among patients. For example, some cases of psoriasis were IL17A predominant, whereas others were IL23A predominant, and others were mixed. Similarly, although most cases of AD were IL13 predominant, others were IL4 predominant.
In a follow-up study, RISH was used to measure type 2 (IL4, IL13), type 1 (IFNG), type 3 (IL17A and IL17F), and Th22 (IL22) cytokines in patients with eczematous dermatitis treated with dupilumab (Singh et al., 2023). In this retrospective study, cytokines were measured in diagnostic biopsies at baseline, and patterns were correlated with response to dupilumab. We hypothesized that patients with a relatively pure Th2 signal would respond optimally to dupilumab, whereas those with a mixed or alternate polarization might respond suboptimally. Indeed, we found that the best responders to dupilumab had the highest expression of IL13, whereas poor or nonresponders had either low or no IL13 and tended to express relatively higher levels of other cytokines such as IFNG and IL17A.
RISH has certain advantages in its potential application for biomarker analysis in inflammatory dermatoses. First, the technique has high specificity for the mRNA of interest with minimal background staining, making it easy to interpret. It can also be performed on diagnostic biopsies that may have already been obtained; has a rapid turnaround; fits well into standard dermatopathology workflow; and does not require a centralized service, specialized knowledge and/or equipment, or data normalization. In contrast to other approaches, histopathologic analysis is also possible in the event of an unexpected diagnosis.
Limitations of the technique include cost (probes cost roughly as much as an antibody for IHC) and difficulty in analyzing a large cytokine panel for each patient because each cytokine target requires a separate probe and staining procedure. In addition, standardized and validated approaches for scoring cytokine expression need to be developed. For some analytes, detecting mRNA as opposed to protein may be a limitation. Furthermore, RISH requires a biopsy to be performed, which may not always be part of routine clinical care. The biopsy could also result in extra costs for tissue processing and dermatopathological evaluation of the biopsy specimen as well as scarring for the patient.
Single-cell RNA sequencing
Single-cell RNA sequencing (scRNA-seq) allows for transcriptomic analysis of individual cells in tissues. scRNA-seq has been applied to human skin in a variety of disease contexts (Kim et al., 2020a), including inflammatory skin diseases (Cheng et al., 2018; He et al., 2020b; Hughes et al., 2020; Kim et al., 2020b; Reynolds et al., 2021; Rojahn et al., 2020).
Recently, Liu et al., 2022b tested the hypothesis that scRNA-seq profiling could classify CIRs. They profiled CD45+ immune cells from the skin of 31 patients, including seven patients with AD, eight patients with psoriasis, two patients with lichen planus, one patient with bullous pemphigoid, six patients with CIRs, and seven healthy controls. They identified AD and psoriasis-specific gene sets within the tissue-resident memory T-cell population, which included Th2- and Th17-associated genes. When they placed CIRs onto this Th2/Th17 map, they found that the classification of CIRs as more AD like correlated with positive response to dupilumab. They created a free web-based tool RashX for users to upload scRNA-seq to assist with molecular classification.
One advantage of scRNA-seq profiling is that it could allow for both treatment selection and the discovery of new biomarkers in inflammatory skin conditions. The creation of centralized databases such as RashX allows for standardization of data analysis and enhanced data sharing.
Limitations of scRNAseq profiling include the high cost and difficulty of integrating scRNA-seq into clinical work-flows. scRNA-seq requires specialized equipment, samples may need to ideally be processed fresh, and this approach may only be possible in academic settings. Tissue dissociation methods also vary and a biopsy is required which leads to scarring. Depending on the tissue processing approach, the entire tissue might be depleted for scRNA-seq and thus histopathologic analysis could require obtaining a second specimen.
Other technologies
Suction blistering is another technique for RNA and proteomic profiling of the skin. Blisters are induced at the dermo–epidermal junction over a few hours. Blister fluid can be used directly for proteomic analysis (Müller et al., 2012; Szegedi et al., 2015), or cells can be collected from the fluid and/or blister roof for transcriptomic analysis. Rojahn et al. (2020) performed scRNA-seq of blister samples from patients with AD and healthy controls and found that the expression profiles of cells from the blister roof recapitulated cell type–specific expression patterns from biopsy samples. Suction blistering was also used to study the persistence of inflammatory signatures in the skin of patients with AD on long-term treatment with dupilumab (Bangert et al., 2021). This method typically does not result in significant scarring. A long sample collection time is a limitation.
RNA can also be detected from skin surface lipids collected noninvasively by wiping the face or scalp with oil-blotting film (Inoue et al., 2022; Shima et al., 2022; Yamamoto-Hanada et al., 2023). Using this method, researchers have shown the capturing of RNA derived from sebaceous glands, epidermis, and hair follicles and showed an increase in inflammation-related RNA molecules in skin surface lipids of patients with AD.
Another method to sample biological material from the skin involves treating the skin with chemical reagents in combination with gentle mechanical abrasion or ultrasound energy (Hwang et al., 2013, 2012; Muradova et al., 2021; Paliwal et al., 2010). Researchers have applied this technique to measure skin surface molecules in AD and psoriasis by embedding a capture-antibody microarray onto a skin patch with quantification by spot ELISA (Røpke et al., 2021; Schaap et al., 2021).
Skin microdialysis has been used to sample soluble materials from the skin of patients with AD (Neisius et al., 2002; Papoiu et al., 2011; Steinhoff et al., 2003) and psoriasis (Buerger et al., 2012; Krogstad et al., 1997; Salgo et al., 2011; Sjögren et al., 2012). This technique may be particularly useful for quantification of drug concentrations (Garcia Ortiz et al., 2009; Neisius et al., 2002; Quist et al., 2016a, 2016b; Turpeinen et al., 1988), continuous monitoring of soluble material concentrations (Clough et al., 2007), or simultaneous sampling and delivery of a therapeutic (Rukwied et al., 2000). This technique is limited in its ability to detect relatively higher-molecular-weight soluble factors such as cytokines (Baumann et al., 2019).
Integration of Biomarkers
Studies in psoriasis have integrated transcriptomic and proteomic profiling to identify molecules that are differentially expressed at both the RNA and protein levels (Piruzian et al., 2010; Swindell et al., 2015). One study also compared gene expression signatures from skin and blood with those of other skin diseases, finding that the blood signature was more psoriasis specific (Swindell et al., 2016). Ungar et al. (2017) combined serum protein measurements with gene expression measurements from lesional and nonlesional skin of patients with AD. Surprisingly, they found that nonlesional skin biomarkers correlated more strongly with serum biomarkers than lesional skin biomarkers. In a pilot study, Foulkes et al. (2019) performed an integrated analysis of serum biomarkers (protein, mRNA, and microRNA) with mRNA measurements from lesional and nonlesional skin to examine the response of patients with psoriasis to TNFα inhibition. The authors built random forest models to classify responses to treatment on the basis of baseline blood and skin measurements. Although underpowered, their pilot study suggests that baseline skin and blood biomarkers may be able to predict response to therapy, which will be explored in future larger studies (Griffiths et al., 2015).
CONCLUSION
An ideal method for biomarker-driven diagnosis and treatment selection is accurate at characterizing the molecular changes present in a patient sample, reproducible across laboratories and conditions, practical in terms of the cost of reagents and analysis, and clinically feasible. Although there are currently no widely used or approved biomarker-based methods to inform therapy selection in AD and psoriasis, several methods are being developed and evaluated. These technologies have limitations, and there are still many open questions (Table 2). We also lack large-scale data at this time showing the clinical and/or financial feasibility of these approaches. Conceptually, given the growing number of biologics with diverse targets for psoriasis, AD, and other inflammatory dermatoses, further development of technologies for biomarker-based therapy selection will be valuable for patients, physicians, and the healthcare system.
Box 1. Open Questions and Limitations of Existing Technologies.
Open Questions:
- How does sampling affect biomarker detection?
- Anatomic location
- Stage of individual lesion
- Duration of underlying disease
- Treatment status of patient
- Severity of disease at moment in time (flare status)
- Technical sampling differences between operators
- What are practical limitations to biomarker detection in the clinic?
- Insurance and reimbursement
- Perceived utility by clinicians
- Time taken to collect samples and coordinate downstream analysis
- Standardization among labs
- Resource limitations
- How do immunologic biomarkers impact diagnosis and disease classification?
- Nosologic considerations (cases with overlapping morphology and immunology)
- Clinically and histologically indeterminate rashes
- Limitations of existing technologies
- Lack large-scale data showing the clinical and/or financial feasibility of these approaches
- Antidrug antibodies are not assessed
- Dose- and patient-specific pharmacokinetics may influence response to biologic therapy (Tsakok et al. 2019; Wilkinson et al. 2019)
ACKNOWLEDGMENTS
We regret that not all relevant works could be included because of space limitations.
Abbreviations:
- AD
atopic dermatitis
- CIR
clinically/histopathologically indeterminate rash
- DBP
dermal biomarker patch
- EASI
Eczema Area and Severity Index
- IF
immunofluorescence
- IHC
immunohistochemistry
- NOS2
nitric oxide synthase 2
- PPV
positive predictive value
- RISH
RNA in situ hybridization
- scRNA-seq
single-cell RNA sequencing
- Th
T helper
Footnotes
CONFLICT OF INTEREST
JMC serves on a data and safety monitoring board for Advarra. WED reports research support from Pfizer, Advanced Cell Diagnostics/Bio-Techne, AbbVie, Incyte, and Bristol Myers Squibb; consulting fees from Eli Lilly, Pfizer, TWI Biotechnology, Incyte, Epiarx Diagnostics, and Bristol Myers Squibb; and licensing fees from EMD/Millipore/Sigma. WED has filed a patent application on the use of cytokine RNA in situ hybridization for personalized diagnosis and treatment selection in inflammatory skin diseases.
REFERENCES
- Aggarwal P, Bowers NL, Muddasani S, Fleischer AB, Feldman SR. Atopic dermatitis and psoriasis are diagnosed clinically and treated empirically. Dermatol Ther (Heidelb) 2022;12:611–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainali C, Valeyev N, Perera G, Williams A, Gudjonsson JE, Ouzounis CA, et al. Transcriptome classification reveals molecular subtypes in psoriasis. BMC Genomics 2012;13:472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akdis M, Akdis CA, Weigl L, Disch R, Blaser K. Skin-homing, CLA+ memory T cells are activated in atopic dermatitis and regulate IgE by an IL-13-dominated cytokine pattern: IgG4 counter-regulation by CLA-memory T cells. J Immunol 1997;159:4611–9. [PubMed] [Google Scholar]
- Andres-Ejarque R, Ale HB, Grys K, Tosi I, Solanky S, Ainali C, et al. Enhanced NF-kB signaling in type-2 dendritic cells at baseline predicts non-response to adalimumab in psoriasis [published correction appears in Nat Commun 2021;12:7358] Nat Commun 2021;12:4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong AW, Puig L, Joshi A, Skup M, Williams D, Li J, et al. Comparison of biologics and oral treatments for plaque psoriasis: A meta-analysis. JAMA Dermatol 2020;156:258–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin LM, Ozawa M, Kikuchi T, Walters IB, Krueger JG. The Majority of Epidermal T cells in psoriasis vulgaris Lesions can Produce Type 1 cytokines, interferon-γ, interleukin-2, and tumor necrosis factor-α, Defining TC1 (cytotoxic T lymphocyte) and TH1 Effector Populations:1 a Type 1 Differentiation Bias is also Measured in Circulating Blood T cells in Psoriatic Patients. J Invest Dermatol 1999;113:752–9. [DOI] [PubMed] [Google Scholar]
- Bagel J, Wang Y, Montgomery P III, Abaya C, Andrade E, Boyce C, et al. A machine learning-based test for predicting response to psoriasis biologics. J Cutan Med Surg 2021;5:621–38. [Google Scholar]
- Bakker DS, de Graaf M, Nierkens S, Delemarre EM, Knol E, van Wijk F, et al. Unraveling heterogeneity in pediatric atopic dermatitis: identification of serum biomarker based patient clusters. J Allergy Clin Immunol 2022;149:125–34. [DOI] [PubMed] [Google Scholar]
- Bangert C, Rindler K, Krausgruber T, Alkon N, Thaler FM, Kurz H, et al. Persistence of mature dendritic cells, TH2A, and Tc2 cells characterize clinically resolved atopic dermatitis under IL-4Rα blockade. Sci Immunol 2021;6(6):eabe2749. [DOI] [PubMed] [Google Scholar]
- Bashir SJ, Chew AL, Anigbogu A, Dreher F, Maibach HI. Physical and physiological effects of stratum corneum tape stripping. Skin Res Technol 2001;7:40–8. [DOI] [PubMed] [Google Scholar]
- Baumann KY, Church MK, Clough GF, Quist SR, Schmelz M, Skov PS, et al. Skin microdialysis: methods, applications and future opportunities-an EAACI position paper. Clin Transl Allergy 2019;9:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck LA, Thaçi D, Hamilton JD, Graham NM, Bieber T, Rocklin R, et al. Dupilumab treatment in adults with moderate-to-severe atopic dermatitis. N Engl J Med 2014;371:130–9. [DOI] [PubMed] [Google Scholar]
- Benson NR, Papenfuss J, Wong R, Motaal A, Tran V, Panko J, et al. An analysis of select pathogenic messages in lesional and non-lesional psoriatic skin using non-invasive tape harvesting. J Invest Dermatol 2006;126:2234–41. [DOI] [PubMed] [Google Scholar]
- Berekméri A, Latzko A, Alase A, Macleod T, Ainscough JS, Laws P, et al. Detection of IL-36γ through noninvasive tape stripping reliably discriminates psoriasis from atopic eczema. J Allergy Clin Immunol 2018;142:988–91.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berekméri A, Tiganescu A, Alase AA, Vital E, Stacey M, Wittmann M. Non-invasive approaches for the diagnosis of autoimmune/autoinflammatory skin diseases—a focus on psoriasis and lupus erythematosus. Front Immunol 2019;10:1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner PM, Israel A, Zhang N, Leonard A, Wen HC, Huynh T, et al. Early-onset pediatric atopic dermatitis is characterized by TH2/TH17/TH22-centered inflammation and lipid alterations. J Allergy Clin Immunol 2018;141:2094–106. [DOI] [PubMed] [Google Scholar]
- Brunner PM, Suárez-Fariñas M, He H, Malik K, Wen HC, Gonzalez J, et al. The atopic dermatitis blood signature is characterized by increases in inflammatory and cardiovascular risk proteins [published correction appears in Sci Rep 2018;8:8439] Sci Rep 2017;7:8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerger C, Richter B, Woth K, Salgo R, Malisiewicz B, Diehl S, et al. Interleukin-1b interferes with epidermal homeostasis through induction of insulin resistance: implications for psoriasis pathogenesis. J Invest Dermatol 2012;132:2206–14. [DOI] [PubMed] [Google Scholar]
- Carlén LM, Sánchez F, Bergman AC, Becker S, Hirschberg D, Franzén B, et al. Proteome analysis of skin distinguishes acute guttate from chronic plaque psoriasis. J Invest Dermatol 2005;124:63–9. [DOI] [PubMed] [Google Scholar]
- Chan JR, Blumenschein W, Murphy E, Diveu C, Wiekowski M, Abbondanzo S, et al. IL-23 stimulates epidermal hyperplasia via TNF and IL-20R2–dependent mechanisms with implications for psoriasis pathogenesis. J Exp Med 2006;203:2577–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JC, Smith LR, Froning KJ, Schwabe BJ, Laxer JA, Caralli LL, et al. CD8+ T cells in psoriatic lesions preferentially use T-cell receptor V beta 3 and/or V beta 13.1 genes. Proceedings of the National Academy of Sciences. Proc Natl Acad Sci USA 1994;91:9282–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JS, Murphy MJ, Singh K, Wang A, Chow RD, Kim SR, et al. IL17A mRNA expression distinguishes palmoplantar psoriasis from hyperkeratotic palmoplantar eczema and mycosis fungoides palmaris et plantaris. JID innovations. 2023. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JB, Sedgewick AJ, Finnegan AI, Harirchian P, Lee J, Kwon S, et al. Transcriptional programming of normal and inflamed human epidermis at single-cell resolution. Cell Rep 2018;25:871–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung PF-Y, Wong CK, Ho AW-Y, Hu S, Chen DP, Lam CW-K. Activationof human eosinophils and epidermal keratinocytes by Th2 cytokine IL-31: implication for the immunopathogenesis of atopic dermatitis. Int Immunol 2010;22:453–67. [DOI] [PubMed] [Google Scholar]
- Choy DF, Hsu DK, Seshasayee D, Fung MAMZ, Martin F, et al. Comparative transcriptomic analyses of atopic dermatitis and psoriasis reveal shared neutrophilic inflammation. J Allergy Clin Immunol 2012;130:1335–43.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen ML, Kezic S, Olesen CM, Agner T. Cytokine concentration across the stratum corneum in atopic dermatitis and healthy controls. Sci Rep 2020;10:21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough GF, Jackson CL, Lee JJP, Jamal SC, Church MK. What can microdialysis tell us about the temporal and spatial generation of cytokines in allergen-induced responses in human skin in vivo? J Invest Dermatol 2007;127:2799–806. [DOI] [PubMed] [Google Scholar]
- Cohen JN, Bowman S, Laszik ZG, North JP. Clinicopathologic overlap of psoriasis, eczema, and psoriasiform dermatoses: a retrospective study of T helper type 2 and 17 subsets, interleukin 36, and β-defensin 2 in spongiotic psoriasiform dermatitis, sebopsoriasis, and tumor necrosis factor α inhibitor–associated dermatitis. J Am Acad Dermatol 2020;82:430–9. [DOI] [PubMed] [Google Scholar]
- Corbett M, Ramessur R, Marshall D, Acencio ML, Ostaszewski M, Barbosa IA, et al. Biomarkers of systemic treatment response in people with psoriasis: a scoping review. Br J Dermatol 2022;187:494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnowicki T, He H, Krueger JG, Guttman-Yassky E. Atopic dermatitis endotypes and implications for targeted therapeutics. J Allergy Clin Immunol 2019;143:11. [DOI] [PubMed] [Google Scholar]
- D’Erme AM, Wilsmann-Theis D, Wagenpfeil J, Hölzel M, Ferring-Schmitt S, Sternberg S, et al. IL-36γ (IL-1F9) is a biomarker for psoriasis skin lesions. J Invest Dermatol 2015;135:1025–32. [DOI] [PubMed] [Google Scholar]
- Di Cesare A, Di Meglio P, Nestle FO. The IL-23/Th17 axis in the immunopathogenesis of psoriasis. J Invest Dermatol 2009;129:1339–50. [DOI] [PubMed] [Google Scholar]
- Dillon SR, Sprecher C, Hammond A, Bilsborough J, Rosenfeld-Franklin M, Presnell SR, et al. Interleukin 31, a cytokine produced by activated T cells, induces dermatitis in mice [published correction appears in Nat Immunol 2005;6:114] Nat Immunol 2004;5:752–60. [DOI] [PubMed] [Google Scholar]
- Dyjack N, Goleva E, Rios C, Kim BE, Bin L, Taylor P, et al. Minimally invasive skin tape strip RNA sequencing identifies novel characteristics of the type 2-high atopic dermatitis disease endotype. J Allergy Clin Immunol 2018;141:1298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esaki H, Brunner PM, Renert-Yuval Y, Czarnowicki T, Huynh T, Tran G, et al. Early-onset pediatric atopic dermatitis is TH2 but also TH17 polarized in skin. J Allergy Clin Immunol 2016;138:1639–51. [DOI] [PubMed] [Google Scholar]
- Foulkes AC, Watson DS, Carr DF, Kenny JG, Slidel T, Parslew R, et al. A framework for multi-omic prediction of treatment response to biologic therapy for psoriasis. J Invest Dermatol 2019;139:100–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Ortiz P, Hansen SH, Shah VP, Menné T, Benfeldt E. Impact of adult atopic dermatitis on topical drug penetration: assessment by cutaneous microdialysis and tape stripping. Acta Derm Venereol 2009;89:33–8. [DOI] [PubMed] [Google Scholar]
- Garzorz-Stark N, Krause L, Lauffer F, Atenhan A, Thomas J, Stark SP, et al. A novel molecular disease classifier for psoriasis and eczema. Exp Dermatol 2016;25:767–74. [DOI] [PubMed] [Google Scholar]
- Gedebjerg A, Johansen C, Kragballe K, Iversen L. IL-20, IL-21 and p40: potential biomarkers of treatment response for ustekinumab. Acta Derm Venereol 2013;93:150–5. [DOI] [PubMed] [Google Scholar]
- Gelato F, Mastorino L, Stepkina E, Cavaliere G, Ribero S, Quaglino P, et al. Is dupilumab as effective in intrinsic atopic dermatitis as it is in extrinsic atopic dermatitis? J Clin Med 2023;12(12):2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittler JK, Shemer A, Suárez-Fariñas M, Fuentes-Duculan J, Gulewicz KJ, Wang CQF, et al. Progressive activation of T(H)2/T(H)22 cytokines and selective epidermal proteins characterizes acute and chronic atopic dermatitis. J Allergy Clin Immunol 2012;130:1344–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goleva E, Calatroni A, LeBeau P, Berdyshev E, Taylor P, Kreimer S, et al. Skin tape proteomics identifies pathways associated with transepidermal water loss and allergen polysensitization in atopic dermatitis. J Allergy Clin Immunol 2020;146:1367–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths CEM, Barnes MR, Burden AD, Nestle FO, Reynolds NJ, Smith CH, et al. Establishing an academic–industrial stratified medicine consortium: psoriasis stratification to optimize relevant therapy. J Invest Dermatol 2015;135:2903–7. [DOI] [PubMed] [Google Scholar]
- Gudjonsson JE, Ding J, Johnston A, Tejasvi T, Guzman AM, Nair RP, et al. Assessment of the psoriatic transcriptome in a large sample: additional regulated genes and comparisons with in vitro models. J Invest Dermatol 2010;130:1829–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman-Yassky E, Bissonnette R, Ungar B, Suárez-Fariñas M, Ardeleanu M, Esaki H, et al. Dupilumab progressively improves systemic and cutaneous abnormalities in patients with atopic dermatitis. J Allergy Clin Immunol 2019a;143:155–72. [DOI] [PubMed] [Google Scholar]
- Guttman-Yassky E, Diaz A, Pavel AB, Fernandes M, Lefferdink R, Erickson T, et al. Use of tape strips to detect immune and barrier abnormalities in the skin of children with early-onset atopic dermatitis. JAMA Dermatol 2019b;155:1358–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman-Yassky E, Nograles KE, Krueger JG. Contrasting pathogenesis of atopic dermatitis and psoriasis—Part I: Clinical and pathologic concepts. J Allergy Clin Immunol 2011a;127:1110–8. [DOI] [PubMed] [Google Scholar]
- Guttman-Yassky E, Nograles KE, Krueger JG. Contrasting pathogenesis of atopic dermatitis and psoriasis—Part II: Immune cell subsets and therapeutic concepts. J Allergy Clin Immunol 2011b;127:1420–32. [DOI] [PubMed] [Google Scholar]
- Guttman-Yassky E, Suárez-Fariñas M, Chiricozzi A, Nograles KE, Shemer A, Fuentes-Duculan J, et al. Broad defects in epidermal cornification in atopic dermatitis identified through genomic analysis. J Allergy Clin Immunol 2009;124:1235–44.e58. [DOI] [PubMed] [Google Scholar]
- He H, Bissonnette R, Wu J, Diaz A, Saint-Cyr Proulx E, Maari C, et al. Tape strips detect distinct immune and barrier profiles in atopic dermatitis and psoriasis. J Allergy Clin Immunol 2021;147:199–212. [DOI] [PubMed] [Google Scholar]
- He H, Olesen CM, Pavel AB, Clausen ML, Wu J, Estrada Y, et al. Tape-Strip proteomic profiling of atopic dermatitis on dupilumab identifies minimally invasive biomarkers. Front Immunol 2020a;11:1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Suryawanshi H, Morozov P, Gay-Mimbrera J, Del Duca E, Kim HJ, et al. Single-cell transcriptome analysis of human skin identifies novel fibroblast subpopulation and enrichment of immune subsets in atopic dermatitis. J Allergy Clin Immunol 2020b;145:1615–28. [DOI] [PubMed] [Google Scholar]
- Hughes AJ, Tawfik SS, Baruah KP, O’Toole EA, O’Shaughnessy RFL. Tape strips in dermatology research. Br J Dermatol 2021;185:26–35. [DOI] [PubMed] [Google Scholar]
- Hughes TK, Wadsworth MH, Gierahn TM, Do T, Weiss D, Andrade PR, et al. Second-strand synthesis-based massively parallel scRNA-seq reveals cellular states and molecular features of human inflammatory skin pathologies. Immunity 2020;53:878–94.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulshof L, Hack DP, Hasnoe QCJ, Dontje B, Jakasa I, Riethmüller C, et al. A minimally invasive tool to study immune response and skin barrier in children with atopic dermatitis. Br J Dermatol 2019;180:621–30. [DOI] [PubMed] [Google Scholar]
- Hwang BH, Doshi N, Tsai KY, Mitragotri S. A reagent to facilitate protein recovery from cells and tissues. Drug Deliv Transl Res 2012;2:297–304. [DOI] [PubMed] [Google Scholar]
- Hwang BH, Tsai KY, Mitragotri S. Optimized lysis buffer reagents for solubilization and preservation of proteins from cells and tissues. Drug Deliv Transl Res 2013;3:428–36. [DOI] [PubMed] [Google Scholar]
- Ibrahim SF, Taft BJ, Wang Y, Lee BI, Andrade E, Abaya C, et al. Minimally invasive skin transcriptome extraction using a dermal biomarker patch. Dermatol Ther (Heidelb) 2022;12:1313–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inkeles MS, Scumpia PO, Swindell WR, Lopez D, Teles RMB, Graeber TG, et al. Comparison of molecular signatures from multiple skin diseases identifies mechanisms of immunopathogenesis. J Invest Dermatol 2015;135:151–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Kuwano T, Uehara Y, Yano M, Oya N, Takada N, et al. Non-invasive human skin transcriptome analysis using mRNA in skin surface lipids. Commun Biol 2022;5:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y, Aihara M, Kirino M, Harada I, Komori-Yamaguchi J, Yamaguchi Y, et al. Interleukin-18 is elevated in the horny layer in patients with atopic dermatitis and is associated with Staphylococcus aureus colonization. Br J Dermatol 2011;164:560–7. [DOI] [PubMed] [Google Scholar]
- Johnston A, Guzman AM, Swindell WR, Wang F, Kang S, Gudjonsson JE. Early tissue responses in psoriasis to the antitumour necrosis factor-α biologic etanercept suggest reduced interleukin-17 receptor expression and signalling. Br J Dermatol 2014;171:97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BE, Goleva E, Kim PS, Norquest K, Bronchick C, Taylor P, et al. Side-by-side comparison of skin biopsies and skin tape stripping highlights abnormal stratum corneum in atopic dermatitis. J Invest Dermatol 2019;139:2387–9.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Chung KB, Kim TG. Application of single-cell RNA sequencing on human skin: technical evolution and challenges. J Dermatol Sci 2020a;99:74–81. [DOI] [PubMed] [Google Scholar]
- Kim D, Kobayashi T, Voisin B, Jo JH, Sakamoto K, Jin SP, et al. Targeted therapy guided by single-cell transcriptomic analysis in drug-induced hypersensitivity syndrome: a case report. Nat Med 2020b;26:236–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Tomalin L, Lee J, Fitz LJ, Berstein G, Correa-da Rosa J, et al. Reduction of inflammatory and cardiovascular proteins in the blood of patients with psoriasis: differential responses between tofacitinib and etanercept after 4 weeks of treatment. J Invest Dermatol 2018;138:273–81. [DOI] [PubMed] [Google Scholar]
- Kim SH, Kim JH, Lee SJ, Jung MS, Jeong DH, Lee KH. Minimally invasive skin sampling and transcriptome analysis using microneedles for skin type biomarker research. Skin Res Technol 2022;28:322–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga C, Kabashima K, Shiraishi N, Kobayashi M, Tokura Y. Possible pathogenic role of Th17 cells for atopic dermatitis. J Invest Dermatol 2008;128:2625–30. [DOI] [PubMed] [Google Scholar]
- Kolbinger F, Loesche C, Valentin MA, Jiang X, Cheng Y, Jarvis P, et al. β-Defensin 2 is a responsive biomarker of IL-17A–driven skin pathology in patients with psoriasis. J Allergy Clin Immunol 2017;139:923–32.e8. [DOI] [PubMed] [Google Scholar]
- Krogstad AL, Lönnroth P, Larson G, Wallin BG. Increased interstitial histamine concentration in the psoriatic plaque. J Invest Dermatol 1997;109:632–5. [DOI] [PubMed] [Google Scholar]
- Krueger JG, Wharton KA, Schlitt T, Suprun M, Torene RI, Jiang X, et al. IL-17A inhibition by secukinumab induces early clinical, histopathologic, and molecular resolution of psoriasis. J Allergy Clin Immunol 2019;144:750–63. [DOI] [PubMed] [Google Scholar]
- Lee E, Trepicchio WL, Oestreicher JL, Pittman D, Wang F, Chamian F, et al. Increased expression of interleukin 23 p19 and p40 in lesional skin of patients with psoriasis vulgaris. J Exp Med 2004;199:125–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DYM. Pathogenesis of atopic dermatitis. J Allergy Clin Immunol 1999;104:S99–108. [DOI] [PubMed] [Google Scholar]
- Leung DYM, Calatroni A, Zaramela LS, LeBeau PK, Dyjack N, Brar K, et al. The nonlesional skin surface distinguishes atopic dermatitis with food allergy as a unique endotype. Sci Transl Med 2019;11:eaav2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Tsoi LC, Swindell WR, Gudjonsson JE, Tejasvi T, Johnston A, et al. Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides insights into disease mechanisms. J Invest Dermatol 2014;134:1828–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med 2006;203:2271–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang H, Cook C, Taylor MA, North JP, Hailer A, et al. Defining patient-level molecular heterogeneity in psoriasis vulgaris based on single-cell transcriptomics. Front Immunol 2022a;13:842651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang H, Taylor M, Cook C, Martínez-Berdeja A, North JP, et al. Classification of human chronic inflammatory skin disease based on single-cell immune profiling. Sci Immunol 2022b;7:eabl9165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowes MA, Kikuchi T, Fuentes-Duculan J, Cardinale I, Zaba LC, Haider AS, et al. Psoriasis vulgaris lesions contain discrete populations of Th1 and Th17 T cells. J Invest Dermatol 2008;128:1207–11. [DOI] [PubMed] [Google Scholar]
- McAleer MA, Jakasa I, Hurault G, Sarvari P, McLean WHI, Tanaka RJ, et al. Systemic and stratum corneum biomarkers of severity in infant atopic dermatitis include markers of innate and T helper cell-related immunity and angiogenesis. Br J Dermatol 2019;180:586–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCluskey D, Benzian-Olsson N, Mahil SK, Hassi NK, Wohnhaas CT, APRICOT and PLUM study team, et al. Single-cell analysis implicates TH17-to-TH2 cell plasticity in the pathogenesis of palmoplantar pustulosis. J Allergy Clin Immunol 2022;150:882–93. [DOI] [PubMed] [Google Scholar]
- Méhul B, Laffet G, Séraїdaris A, Russo L, Fogel P, Carlavan I, et al. Noninvasive proteome analysis of psoriatic stratum corneum reflects pathophysiological pathways and is useful for drug profiling. Br J Dermatol 2017;177:470–88. [DOI] [PubMed] [Google Scholar]
- Méhul B, Ménigot C, Fogel P, Seraidaris A, Genette A, Pascual T, et al. Proteomic analysis of stratum corneum in cutaneous T-cell Lymphomas and psoriasis. Exp Dermatol 2019;28:317–21. [DOI] [PubMed] [Google Scholar]
- Mikhaylov D, Del Duca E, Guttman-Yassky E. Proteomic signatures of inflammatory skin diseases: a focus on atopic dermatitis. Expert Rev Proteomics 2021a;18:345–61. [DOI] [PubMed] [Google Scholar]
- Mikhaylov D, Del Duca E, Olesen CM, He H, Wu J, Ungar B, et al. Transcriptomic profiling of tape-strips from moderate to severe atopic dermatitis patients treated with dupilumab. Dermatitis 2021b;32:S71–80. [DOI] [PubMed] [Google Scholar]
- Mindera Health. An exploratory, multicenter, observational study to examine RNA biomarkers of psoriasis subjects through the application of the Mindera kit Part 2 (STAMP-2). https://clinicaltrials.gov/ct2/show/NCT04904315 (accessed April 1, 2023).
- Miranda E, Roberts J, Novick S, Lapointe JM, Bruijnzeel-Koomen C, Thijs J, et al. Immunohistochemical characterization of the IL-13:IL-4 receptor α axis in the skin of adult patients with moderate to severe atopic dermatitis and healthy controls. J Invest Dermatol 2021;141:440–3.e4. [DOI] [PubMed] [Google Scholar]
- Möbus L, Rodriguez E, Harder I, Stölzl D, Boraczynski N, Gerdes S, et al. Atopic dermatitis displays stable and dynamic skin transcriptome signatures. J Allergy Clin Immunol 2021;147:213–23. [DOI] [PubMed] [Google Scholar]
- Morhenn VB, Chang EY, Rheins LA. A noninvasive method for quantifying and distinguishing inflammatory skin reactions. J Am Acad Dermatol 1999;41:687–92. [DOI] [PubMed] [Google Scholar]
- Moy AP, Murali M, Kroshinsky D, Duncan LM, Nazarian RM. Immunologic overlap of helper T-cell Subtypes 17 and 22 in erythrodermic psoriasis and atopic dermatitis. JAMA Dermatol 2015;151:753–60. [DOI] [PubMed] [Google Scholar]
- Müller AC, Breitwieser FP, Fischer H, Schuster C, Brandt O, Colinge J, et al. A comparative proteomic study of human skin suction blister fluid from healthy individuals using immunodepletion and iTRAQ labeling. J Proteome Res 2012;11:3715–27. [DOI] [PubMed] [Google Scholar]
- Muradova E, Patel N, Sell B, Bittencourt BB, Ojeda SS, Adelmann CH, et al. Noninvasive assessment of epidermal genomic markers of UV exposure in skin. J Invest Dermatol 2021;141:124–31.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neisius U, Olsson R, Rukwied R, Lischetzki G, Schmelz M. Prostaglandin E2 induces vasodilation and pruritus, but no protein extravasation in atopic dermatitis and controls. J Am Acad Dermatol 2002;47:28–32. [DOI] [PubMed] [Google Scholar]
- Noda S, Suárez-Fariñas M, Ungar B, Kim SJ, de Guzman Strong C, Xu H, et al. The Asian atopic dermatitis phenotype combines features of atopic dermatitis and psoriasis with increased TH17 polarization. J Allergy Clin Immunol 2015;136:1254–64. [DOI] [PubMed] [Google Scholar]
- Noh S, Jin S, Park CO, Lee YS, Lee N, Lee J, et al. Elevated Galectin-10 expression of IL-22-producing T cells in patients with atopic dermatitis. J Invest Dermatol 2016;136:328–31. [DOI] [PubMed] [Google Scholar]
- Nomura I, Gao B, Boguniewicz M, Darst MA, Travers JB, Leung DYm. Distinct patterns of gene expression in the skin lesions of atopic dermatitis and psoriasis: a gene microarray analysis. J Allergy Clin Immunol 2003;112:1195–202. [DOI] [PubMed] [Google Scholar]
- Oestreicher JL, Walters IB, Kikuchi T, Gilleaudeau P, Surette J, Schwertschlag U, et al. Molecular classification of psoriasis disease-associated genes through pharmacogenomic expression profiling. Pharmacogenomics J 2001;1:272–87. [DOI] [PubMed] [Google Scholar]
- Paliwal S, Ogura M, Mitragotri S. One-step acquisition of functional biomolecules from tissues. Proceedings of the National Academy of Sciences. Proc Natl Acad Sci USA 2010;107:14627–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoiu ADP, Wang H, Nattkemper L, Tey HL, Ishiuji Y, Chan YH, et al. A study of serum concentrations and dermal levels of NGF in atopic dermatitis and healthy subjects. Neuropeptides 2011;45:417–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavel AB, Renert-Yuval Y, Wu J, Del Duca E, Diaz A, Lefferdink R, et al. Tape strips from early-onset pediatric atopic dermatitis highlight disease abnormalities in nonlesional skin. Allergy 2021;76:314–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavel AB, Zhou L, Diaz A, Ungar B, Dan J, He H, et al. The proteomic skin profile of moderate-to-severe atopic dermatitis patients shows an inflammatory signature. J Am Acad Dermatol 2020;82:690–9. [DOI] [PubMed] [Google Scholar]
- Piruzian E, Bruskin S, Ishkin A, Abdeev R, Moshkovskii S, Melnik S, et al. Integrated network analysis of transcriptomic and proteomic data in psoriasis. BMC Syst Biol 2010;4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskin G, Sylva-Steenland RMR, Bos JD, Teunissen MBM. In vitro and in situ expression of IL-23 by keratinocytes in healthy skin and psoriasis lesions: enhanced expression in psoriatic skin. J Immunol 2006;176:1908–15. [DOI] [PubMed] [Google Scholar]
- Pourani MR, Abdollahimajd F, Zargari O, Shahidi Dadras M. Soluble biomarkers for diagnosis, monitoring, and therapeutic response assessment in psoriasis. J Dermatolog Treat 2022;33:1967–74. [DOI] [PubMed] [Google Scholar]
- Quaranta M, Knapp B, Garzorz N, Mattii M, Pullabhatla V, Pennino D, et al. Intraindividual genome expression analysis reveals a specific molecular signature of psoriasis and eczema. Sci Transl Med 2014;6:244ra90. [DOI] [PubMed] [Google Scholar]
- Quick AP, Farberg AS, Goldberg MS, Wilkinson J, Silverberg JI. Feasibility of a novel, non-invasive sample collection technique to develop a molecular test guiding therapeutic selection for patients with atopic dermatitis and psoriasis. https://castlebiosciences.com/wp-content/uploads/2022/04/RAD22_Quick_Poster.pdf; 2022. accessed April 1, 2023.
- Quist SR, Quist J, Birkenmaier J, Stauch T, Gollnick HP. Pharmacokinetic profile of methotrexate in psoriatic skin via the oral or subcutaneous route using dermal microdialysis showing higher methotrexate bioavailability in psoriasis plaques than in non-lesional skin. J Eur Acad Dermatol Venereol 2016a;30:1537–43. [DOI] [PubMed] [Google Scholar]
- Quist SR, Wiswedel I, Doering I, Quist J, Gollnick HP. Effects of topical tacrolimus and polyunsaturated fatty acids on in vivo release of eicosanoids in atopic dermatitis during dermal microdialysis. Acta Derm Venereol 2016b;96:905–9. [DOI] [PubMed] [Google Scholar]
- Ramessur R, Corbett M, Marshall D, Acencio ML, Barbosa IA, Dand N, et al. Biomarkers of disease progression in people with psoriasis: a scoping review. Br J Dermatol 2022;187:481–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renert-Yuval Y, Thyssen JP, Bissonnette R, Bieber T, Kabashima K, Hijnen D, et al. Biomarkers in atopic dermatitis-a review on behalf of the International Eczema Council. J Allergy Clin Immunol 2021;147:1174–90.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renz H, Jujo K, Bradley KL, Domenico J, Gelfand EW, Leung DY. Enhanced IL-4 production and IL-4 receptor expression in atopic dermatitis and their modulation by interferon-gamma. J Invest Dermatol 1992;99:403–8. [DOI] [PubMed] [Google Scholar]
- Reynolds G, Vegh P, Fletcher J, Poyner EFM, Stephenson E, Goh I, et al. Developmental cell programs are co-opted in inflammatory skin disease. Science 2021;371:eaba6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez E, Baurecht H, Wahn AF, Kretschmer A, Hotze M, Zeilinger S, et al. An integrated epigenetic and transcriptomic analysis reveals distinct tissue-specific patterns of DNA methylation associated with atopic dermatitis. J Invest Dermatol 2014;134:1873–83. [DOI] [PubMed] [Google Scholar]
- Rojahn TB, Vorstandlechner V, Krausgruber T, Bauer WM, Alkon N, Bangert C, et al. Single-cell transcriptomics combined with interstitial fluid proteomics defines cell type–specific immune regulation in atopic dermatitis. J Allergy Clin Immunol 2020;146:1056–69. [DOI] [PubMed] [Google Scholar]
- Røpke MA, Mekulova A, Pipper C, Eisen M, Pender K, Spee P, et al. Non-invasive assessment of soluble skin surface biomarkers in atopic dermatitis patients-effect of treatment. Skin Res Technol 2021;27:715–22. [DOI] [PubMed] [Google Scholar]
- Rosa da JC, Kim J, Tian S, Tomalin LE, Krueger JG, Suárez-Fariñas M. Shrinking the psoriasis assessment gap: early gene-expression profiling accurately predicts response to long-term treatment. J invest dermatol 2017;137:305–12. [DOI] [PubMed] [Google Scholar]
- Rukwied R, Lischetzki G, McGlone F, Heyer G, Schmelz M. Mast cell mediators other than histamine induce pruritus in atopic dermatitis patients: a dermal microdialysis study. Br J Dermatol 2000;142:1114–20. [DOI] [PubMed] [Google Scholar]
- Salgo R, Thaçi D, Boehncke S, Diehl S, Hofmann M, Boehncke WH. Microdialysis documents changes in the micromilieu of psoriatic plaques under continuous systemic therapy. Exp Dermatol 2011;20:130–3. [DOI] [PubMed] [Google Scholar]
- Samant PP, Niedzwiecki MM, Raviele N, Tran V, Mena-Lapaix J, Walker DI, et al. Sampling interstitial fluid from human skin using a microneedle patch. Sci Transl Med 2020;12:eaaw0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal RD, Pavel AB, Glickman J, Chan TC, Zheng X, Zhang N, et al. Atopic dermatitis in African American patients is TH2/TH22-skewed with TH1/ TH17 attenuation. Ann Allergy Asthma Immunol 2019;122:99–110.e6. [DOI] [PubMed] [Google Scholar]
- Schaap MJ, Bruins FM, He X, Orro K, Peppelman M, van Erp PEJ, et al. Skin surface protein detection by transdermal analysis patches in pediatric psoriasis. Skin Pharmacol Physiol 2021;34:271–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima K, Inoue T, Uehara Y, Iwamura M, Fukagawa S, Kuwano T, et al. Non-invasive transcriptomic analysis using mRNAs in skin surface lipids obtained from children with mild-to-moderate atopic dermatitis. J Eur Acad Dermatol Venereol 2022;36:1477–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverberg JI, Thyssen JP, Fahrbach K, Mickle K, Cappelleri JC, Romero W, et al. Comparative efficacy and safety of systemic therapies used in moderate-to-severe atopic dermatitis: a systematic literature review and network meta-analysis. J Eur Acad Dermatol Venereol 2021;35:1797–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen S, Brøgger P, Kezic S, Thyssen JP, Skov L. Comparison of cytokines in skin biopsies and tape strips from adults with atopic dermatitis. Dermatology 2021;237:940–5. [DOI] [PubMed] [Google Scholar]
- Singh K, Valido K, Swallow M, Okifo KO, Wang A, Cohen JM, et al. Baseline skin cytokine profiles determined by RNA in situ hybridization correlate with response to dupilumab in patients with eczematous dermatitis. J Am Acad Dermatol 2023;88:1094–100. [DOI] [PubMed] [Google Scholar]
- Sjögren F, Davidsson K, Sjöström M, Anderson CD. Cutaneous microdialysis: cytokine evidence for altered innate reactivity in the skin of psoriasis patients? AAPS J 2012;14:187–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolev VV, Soboleva AG, Denisova EV, Pechatnikova EA, Dvoryankova E, Korsunskaya IM, et al. Proteomic studies of psoriasis. Biomedicines 2022;10:619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofen H, Smith S, Matheson RT, Leonardi CL, Calderon C, Brodmerkel C, et al. Guselkumab (an IL-23–specific mAb) demonstrates clinical and molecular response in patients with moderate-to-severe psoriasis. J Allergy Clin Immunol 2014;133:1032–40. [DOI] [PubMed] [Google Scholar]
- Sølberg JBK, Quaade AS, Jacobsen SB, Andersen JD, Kampmann ML, Morling N, et al. The transcriptome of hand eczema assessed by tape stripping. Contact Dermatitis 2022;86:71–9. [DOI] [PubMed] [Google Scholar]
- Steinhoff M, Neisius U, Ikoma A, Fartasch M, Heyer G, Skov PS, et al. Proteinase-activated receptor-2 mediates itch: a novel pathway for pruritus in human skin. J Neurosci 2003;23:6176–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-Fariñas M, Li K, Fuentes-Duculan J, Hayden K, Brodmerkel C, Krueger JG. Expanding the psoriasis disease profile: interrogation of the skin and serum of patients with moderate-to-severe psoriasis [published correction appears in J Invest Dermatol 2015;135:2901–2902] J Invest Dermatol 2012;132:2552–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-Fariñas M, Ungar B, Correa da Rosa J, Ewald DA, Rozenblit M, Gonzalez J, et al. RNA sequencing atopic dermatitis transcriptome profiling provides insights into novel disease mechanisms with potential therapeutic implications. J Allergy Clin Immunol 2015;135:1218–27. [DOI] [PubMed] [Google Scholar]
- Swindell WR, Johnston A, Voorhees JJ, Elder JT, Gudjonsson JE. Dissecting the psoriasis transcriptome: inflammatory- and cytokine-driven gene expression in lesions from 163 patients. BMC Genomics 2013;14:527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR, Remmer HA, Sarkar MK, Xing X, Barnes DH, Wolterink L, et al. Proteogenomic analysis of psoriasis reveals discordant and concordant changes in mRNA and protein abundance. Genome Med 2015;7:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR, Sarkar MK, Liang Y, Xing X, Gudjonsson JE. Cross-disease transcriptomics: unique IL-17A signaling in psoriasis lesions and an autoimmune PBMC signature. J Invest Dermatol 2016;136:1820–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR, Xing X, Stuart PE, Chen CS, Aphale A, Nair RP, et al. Heterogeneity of inflammatory and cytokine networks in chronic plaque psoriasis. PLoS One 2012;7:e34594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szegedi K, Lutter R, Res PC, Bos JD, Luiten RM, Kezic S, et al. Cytokine profiles in interstitial fluid from chronic atopic dermatitis skin. J Eur Acad Dermatol Venereol 2015;29:2136–44. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Arai I, Sugimoto M, Honma Y, Futaki N, Nakamura A, et al. Involvement of IL-31 on scratching behavior in NC/Nga mice with atopic-like dermatitis. Exp Dermatol 2006;15:161–7. [DOI] [PubMed] [Google Scholar]
- Teunissen MB, Koomen CW, de Waal Malefyt R, Wierenga EA, Bos JD. Interleukin-17 and interferon-γ synergize in the enhancement of proinflammatory cytokine production by human keratinocytes. J Invest Dermatol 1998;111:645–9. [DOI] [PubMed] [Google Scholar]
- Thijs JL, Strickland I, Bruijnzeel-Koomen CAFM, Nierkens S, Giovannone B, Csomor E, et al. Moving toward endotypes in atopic dermatitis: identification of patient clusters based on serum biomarker analysis [published correction appears in J Allergy Clin Immunol 2018;142:714. J Allergy Clin Immunol 2017;140:730–7. [DOI] [PubMed] [Google Scholar]
- Tsakok T, Wilson N, Dand N, Loeff FC, Bloem K, Baudry D, et al. Association of serum ustekinumab levels with clinical response in psoriasis. JAMA Dermatol 2019;155:1235–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi LC, Iyer MK, Stuart PE, Swindell WR, Gudjonsson JE, Tejasvi T, et al. Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol 2015;16:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi LC, Rodriguez E, Degenhardt F, Baurecht H, Wehkamp U, Volks N, et al. Atopic dermatitis is an IL-13–dominant disease with greater molecular heterogeneity compared to psoriasis. J Invest Dermatol 2019;139:1480–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi LC, Xing X, Xing E, Wasikowski R, Shao S, Zeng C, et al. Noninvasive tape-stripping with high-resolution RNA profiling effectively captures a Preinflammatory State in nonlesional psoriatic skin. J Invest Dermatol 2022;142:1587–96.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turpeinen M, Mashkilleyson N, Björkstén F, Salo OP. Percutaneous absorption of hydrocortisone during exacerbation and remission of atopic dermatitis in adults. Acta Derm Venereol 1988;68:331–5. [PubMed] [Google Scholar]
- Ungar B, Garcet S, Gonzalez J, Dhingra N, Correa da Rosa J, Shemer A, et al. An integrated model of atopic dermatitis biomarkers highlights the systemic nature of the disease. J Invest Dermatol 2017;137:603–13. [DOI] [PubMed] [Google Scholar]
- van der Heijden FL, Wierenga EA, Bos JD, Kapsenberg ML. High frequency of IL-4–producing CD4+ allergen-specific T lymphocytes in atopic dermatitis lesional skin. J Invest Dermatol 1991;97:389–94. [DOI] [PubMed] [Google Scholar]
- van Reijsen FC, Bruijnzeel-Koomen CAFM, Kalthoff FS, Maggi E, Romagnani S, Westland JKT, et al. Skin-derived aeroallergen-specific T-cell clones of Th2 phenotype in patients with atopic dermatitis. J Allergy Clin Immunol 1992;90:184–93. [DOI] [PubMed] [Google Scholar]
- Wang A, Fogel AL, Murphy MJ, Panse G, McGeary MK, McNiff JM, et al. Cytokine RNA in situ hybridization permits individualized molecular phenotyping in biopsies of psoriasis and atopic dermatitis. JID Innov 2021a;1:100021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 2012;14:22–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Suárez-Fariñas M, Estrada Y, Parker ML, Greenlees L, Stephens G, et al. Identification of unique proteomic signatures in allergic and non-allergic skin disease. Clin Exp Allergy 2017;47:1456–67. [DOI] [PubMed] [Google Scholar]
- Wang Z, Luan J, Seth A, Liu L, You M, Gupta P, et al. Microneedle patch for the ultrasensitive quantification of protein biomarkers in interstitial fluid. Nat Biomed Eng 2021b;5:64–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson N, Tsakok T, Dand N, Bloem K, Duckworth M, Baudry D, et al. Defining the therapeutic range for adalimumab and predicting response in psoriasis: A multicenter prospective observational cohort study. J Invest Dermatol 2019;139:115–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong R, Tran V, Morhenn V, Hung SP, Andersen B, Ito E, et al. Use of RT-PCR and DNA microarrays to characterize RNA recovered by non-invasive tape harvesting of normal and inflamed skin. J Invest Dermatol 2004;123:159–67. [DOI] [PubMed] [Google Scholar]
- Wongvibulsin S, Sutaria N, Kannan S, Alphonse MP, Belzberg M, Williams KA, et al. Transcriptomic analysis of atopic dermatitis in African Americans is characterized by Th2/Th17-centered cutaneous immune activation. Sci Rep 2021;11:11175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Gu C, Wang S, Yin H, Qiu Z, Luo Y, et al. Serum biomarker-based endotypes of atopic dermatitis in China and prediction for efficacy of dupilumab. Br J Dermatol 2023;188:649–60. [DOI] [PubMed] [Google Scholar]
- Yamamoto-Hanada K, Saito-Abe M, Shima K, Fukagawa S, Uehara Y, Ueda Y, et al. mRNAs in skin surface lipids unveiled atopic dermatitis at 1 month [epub ahead of print]. J Eur Acad Dermatol Venereol 2023. 10.1111/jdv.19017 (accessed March 1, 2023). [DOI] [PubMed] [Google Scholar]
- Zhou X, Krueger JG, Kao MC, Lee E, Du F, Menter A, et al. Novel mechanisms of T-cell and dendritic cell activation revealed by profiling of psoriasis on the 63,100-element oligonucleotide array. Physiol Genomics 2003;13:69–78. [DOI] [PubMed] [Google Scholar]


