Figure 2. Skin sampling and processing for different investigational techniques for personalized therapy selection in inflammatory skin disease.
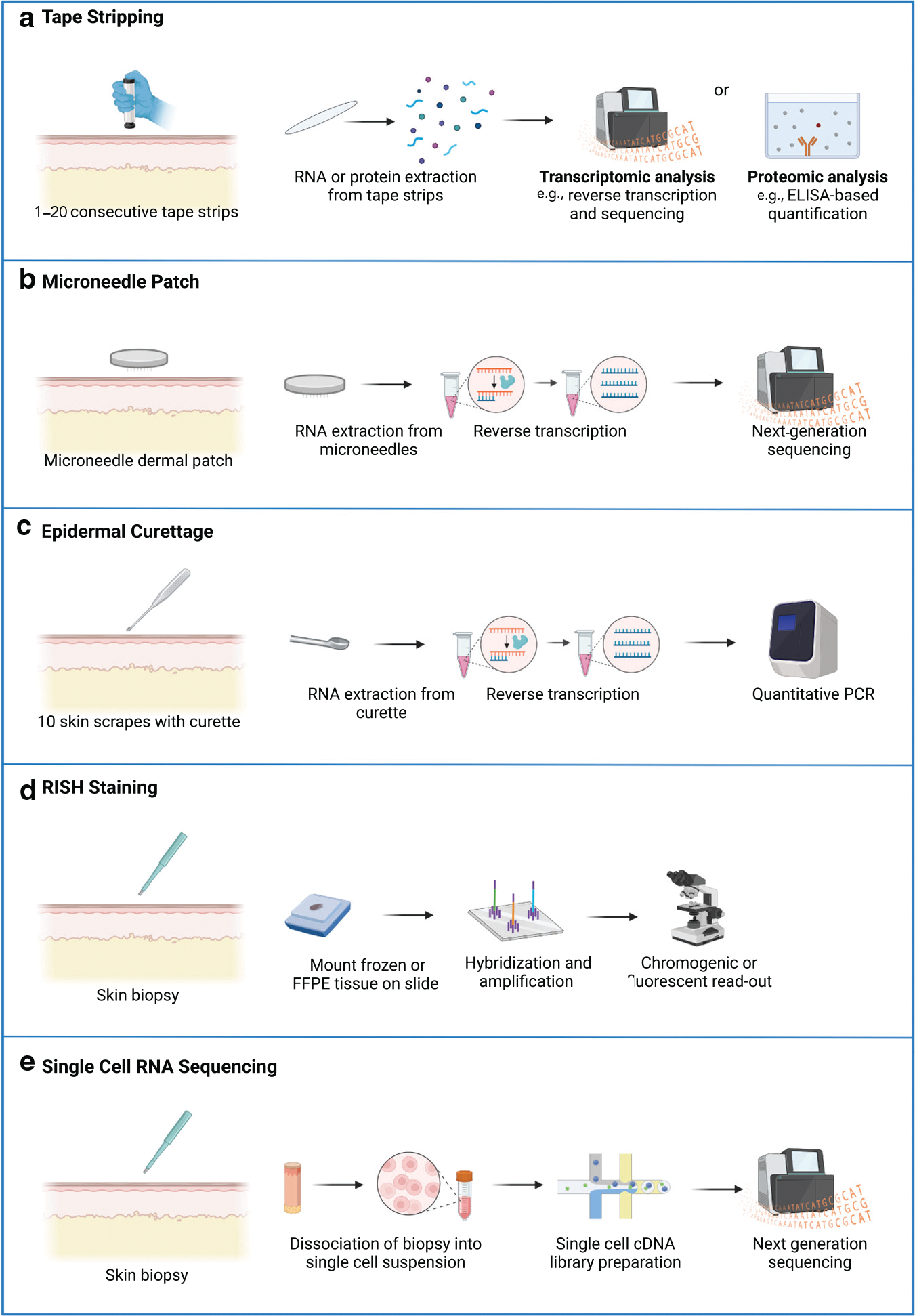
(a) For tape stripping, an applicator is applied to the skin for consecutive tape strips. RNA or proteins are extracted from the tape strips and quantified. (b) For the microneedle-based dermal biomarker patch (Mindera Health, San Diego, CA), a microneedle patch which has probes extending into the superficial dermis is applied to the skin. RNA is extracted, reverse transcribed, and sequenced. (c) For molecular profiling from epidermal curettage (Castle Biosciences, Friendswood, TX), the skin is scraped with a curette, and RNA is isolated from the skin sample and analyzed by RT-PCR. (d) For RISH, a skin biopsy is obtained. A probe complementary to the RNA molecule of interest is added to the tissue slide, and a chromogenic or fluorescent marker is added in the detection step. (e) For single-cell RNA sequencing, a biopsy is obtained, and the skin sample is dissociated into single cells. Single-cell cDNA library preparation is performed, and the cDNA library is sequenced using next-generation sequencing. FFPE, formalin-fixed, paraffin-embedded; RISH, RNA in situ hybridization.
