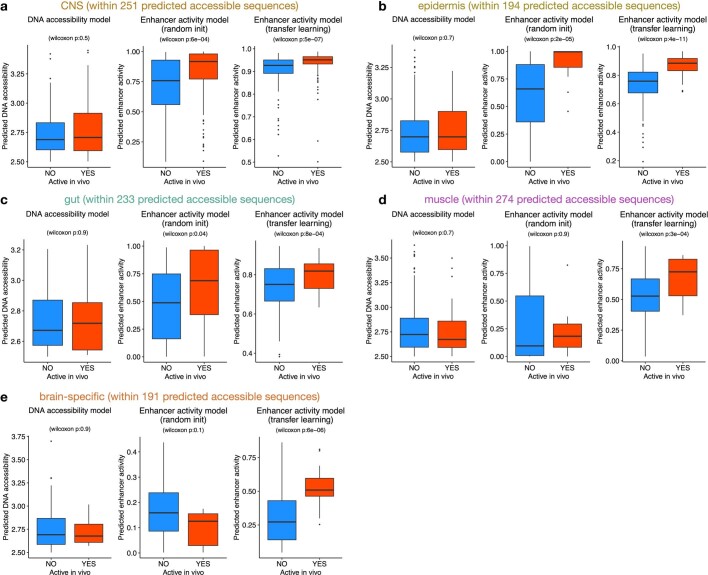
Extended Data Fig. 5. Predictive value of DNA accessibility and enhancer-activity models for predicted accessible sequences.
a-e) For each tissue, sequences in the test set were selected based on a predicted DNA accessibility value higher than 2.5 and scored with the different models (total number of selected sequences shown in panel title). Sequences inactive (blue) or active (red) in vivo are shown in boxplots in function of their scores by the DNA accessibility model, enhancer activity model starting from random initialization, and enhancer activity model using transfer learning. P-values from two-sided Wilcoxon rank-sum test are shown for each comparison between inactive and active sequences. Numbers of predicted accessible sequences used for statistics per tissue: CNS – 251, epidermis – 194, gut – 233, muscle – 274, brain-specific – 191. The boxplots mark the median, upper and lower quartiles and 1.5× interquartile range (whiskers); outliers are shown individually.