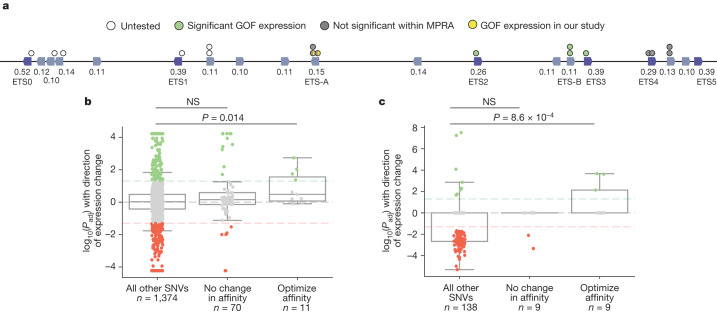
Fig. 5. Affinity-optimizing SNVs drive GOF expression in the ZRS and IFNβ enhanceosome.
a, Schematic of the human ZRS showing 19 putative ETS sites and 17 SNVs that increase the affinity of ETS sites by 1.6-fold or more. ETS sites identified from previous studies (ETS0–ETS5) are labelled15,51. We annotated changes in expression caused by these SNVs on the basis of results from a saturation mutagenesis MPRA and our study. b,c, Box plots showing all SNVs tested within MPRA mutagenesis experiments, their significance and their effects on expression. The bounds of the box plots define the 25th, 50th and 75th percentiles, and whiskers are 1.5× the interquartile range. The y axis is the log of the adjusted P value (Padj) with the direction of expression change (positive values indicate an increase in expression and negative values indicate a decrease in expression). Dashed horizontal lines indicate significance thresholds at P = .05. Each dot represents a tested SNV. SNVs that have a significant increase in expression are shown in green, those with no significant change in expression are grey and those with a significant decrease in expression are red. For each plot, we show SNVs that increase the affinity of the transcription factor, SNVs within the TFBS that do not alter affinity and all other SNVs. b, Analysis of the ETS TFBS in a saturation mutagenesis MPRA on a 485-bp region of the ZRS enhancer. c, Analysis of the IRF TFBS in a saturation mutagenesis MPRA on the IFNβ enhanceosome. We used one-tailed Mann–Whitney U tests to determine any significant enrichment for GOF enhancer activity.