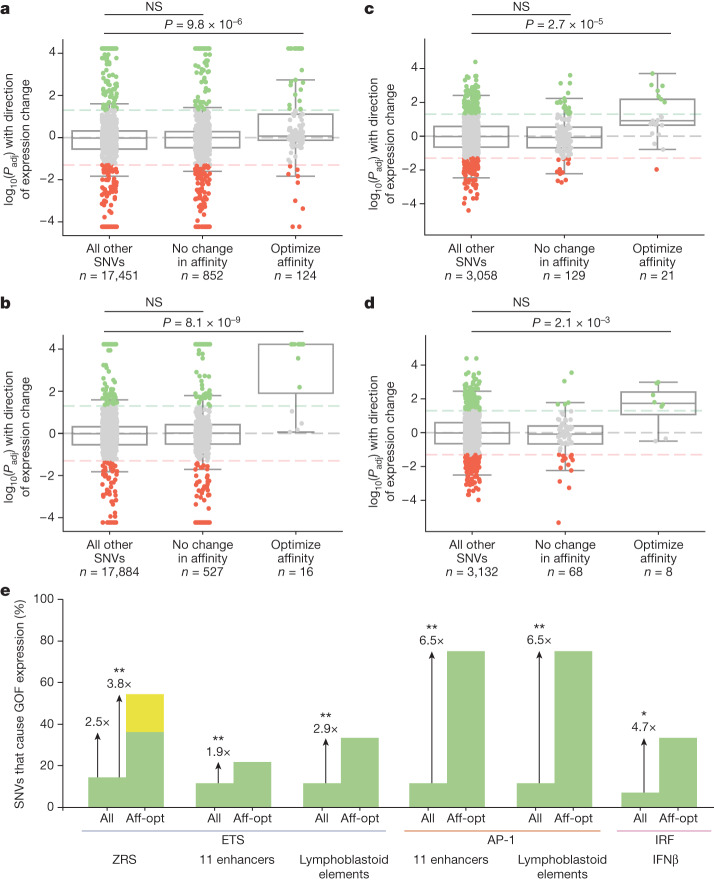
Fig. 6. Affinity-optimizing SNVs drive GOF expression in a wide variety of disease-associated enhancers.
a–d, Analysis of MPRAs for a variety of enhancers in different cell types. Box plots showing all SNVs tested within MPRA mutagenesis experiments and their significance and effects on expression. The bounds of the box plots define the 25th, 50th and 75th percentiles, and whiskers are 1.5× the interquartile range. One-tailed Mann–Whitney U test. a,b, Analysis of saturation mutagenesis MPRA assays of 11 disease-associated enhancers comparing all SNVs, SNVs within TFBSs that do not alter affinity and SNVs that increase the affinity of TFBSs for ETS (a) and AP-1 (b). c,d, Analysis of MPRA comparing the effect of SNVs within lymphoblastoid regulatory elements for the ETS TFBS (c) and the AP-1 TFBS (d). e, Filtering for affinity-optimizing (aff-opt) SNVs significantly increases our ability to predict causal GOF enhancer variants. The bar graph shows the percentage of all SNVs that lead to GOF expression relative to the percentage of affinity-optimizing SNVs that lead to GOF expression. Green bars indicate SNVs that cause GOF expression within analysed MPRA datasets; yellow bar indicates SNVs that cause GOF expression in our the current study—namely, French 2 and Indian 2. Fisher’s exact test was used to determine any significant enrichment for GOF expression in the all and aff-opt categories: **P < 0.001, *P < 0.01. Affinity-optimizing SNVs are those that lead to a fold change of at least 1.6 for ETS because this is the fold change for French 2, Indian 2 and Syn 0.25. Affinity-optimizing SNVs for AP-1 and IRF are those that cause a fold change of at least 1.5.