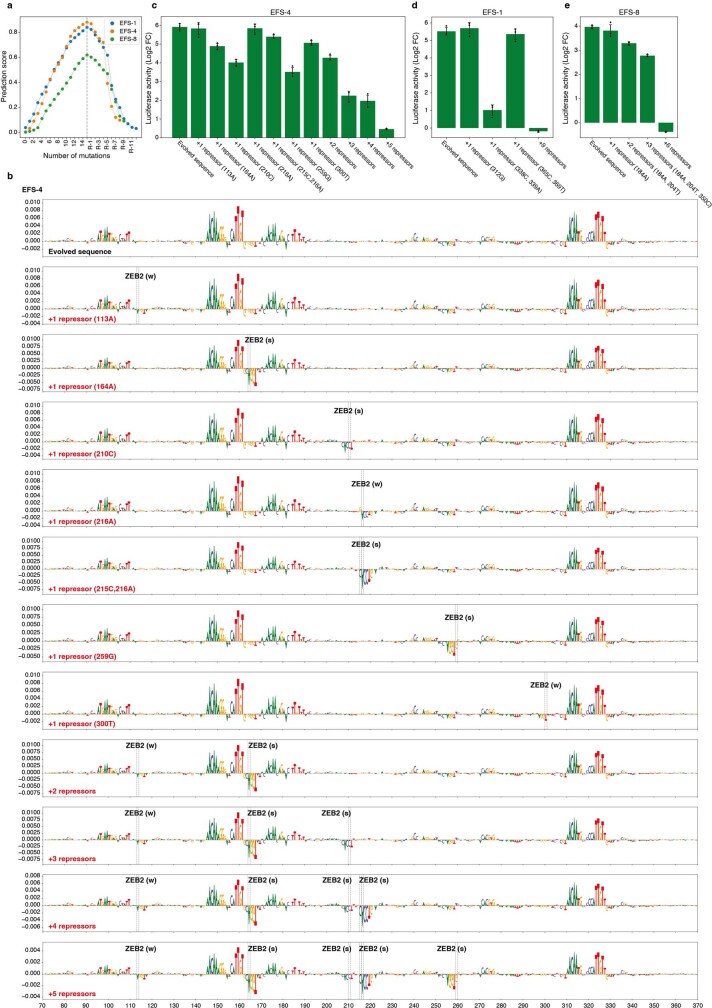
Extended Data Fig. 8. ZEB2 repression of in silico evolved MEL enhancers.
a, Prediction scores for each mutational step and after the addition of repressor sites for 3 EFS sequences. b, Nucleotide contribution scores (DeepMEL2 MEL class) showing the creation of single or multiple repressor binding sites by single or double mutations in the EFS-4 sequence. c–e, In vivo enhancer activity of EFS-4 (c), EFS-1 (d) and EFS-8 (e) after the generation of repressor binding sites. ZEB2 motif annotation is indicated with strong (s) or weak (w) motif instances. The error bars in c–e, show the standard error of the mean (n = 3 biological replicates).