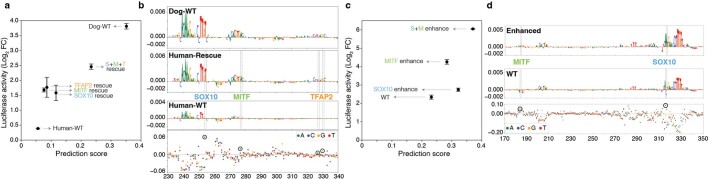
Extended Data Fig. 9. Human enhancer rescue.
In the fly brain, we applied in silico sequence evolution to create enhancers from genomic regions with high scores that did not show chromatin accessibility and could consequently be considered as ‘near-enhancer’ sequences. We extended this approach to MEL enhancers. We started from a human sequence that has no MEL enhancer activity, but its homologous sequence in the dog genome is accessible and active as MEL enhancer. We used DeepMEL to introduce 4 mutations that restored the activator binding sites in the human sequence, resulting in a rescue of the activity, as measured by luciferase activity. a, Dot plot showing the mean luciferase signal (log2 fold-change (FC) over Renilla) versus prediction score for the MEL class of the WT human and dog genomic sequences and the rescued human sequences. b, Nucleotide contribution scores of the dog, human-rescued and human-WT sequences (top 3 rows) and in silico saturation mutagenesis assay of human-WT sequence (bottom). c, As a variation of this approach, we introduced two mutations in a weak MEL enhancer which resulted in a 10-fold increase in enhancer activity. Dot plot showing the mean luciferase signal (log2 FC over Renilla) versus prediction score for the MEL class of the wild-type and enhanced enhancers. d, Nucleotide contribution scores of the wild-type (middle) and enhanced (top) enhancers and in silico saturation mutagenesis assay of wild-type enhancer (bottom). In a, c, the error bars show the standard error of the mean (n = 3 biological replicates). S, SOX10; M, MITF; T, TFAP2. In b,d, each dot on the saturation mutagenesis plot represents a single mutation and its effect on the prediction score (y axis). The position of the mutations is shown with dashed lines and circles.