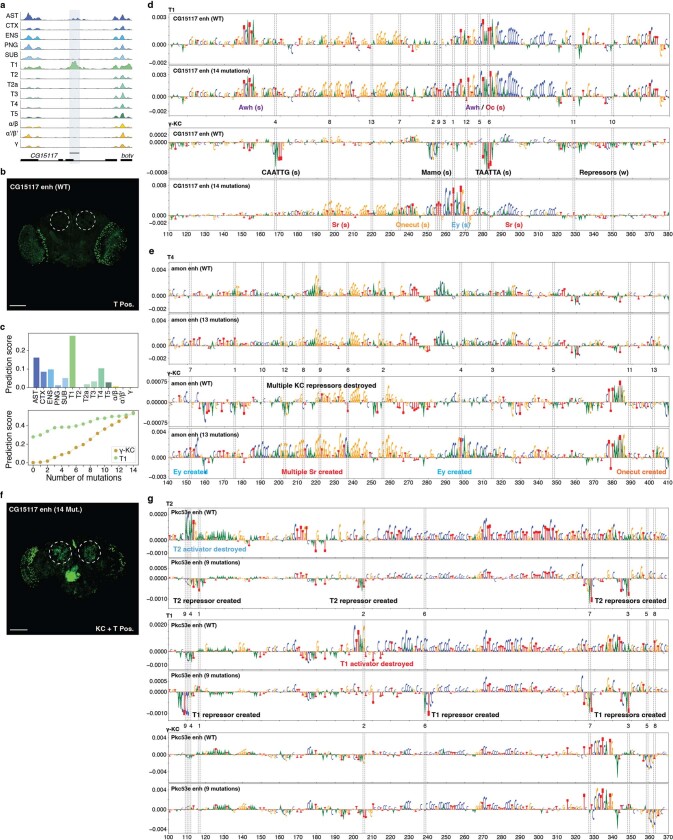
Extended Data Fig. 3. Enhancer design toward multiple cell type codes.
a, Chromatin accessibility profile near CG15117 gene. b, In vivo enhancer activity of the wild-type (WT) CG15117 enhancer. c, CG15117 enhancer prediction scores for each cell type (top) and prediction scores for the γ-KC and T1 classes after each mutational step. d, Nucleotide contribution scores of WT CG15117 enhancer sequence and after 14 mutations for T1 (top) and γ-KC (bottom). e, Nucleotide contribution scores of WT amon enhancer sequence and after 13 mutations for T4 (top) and γ-KC (bottom). f, In vivo enhancer activity of the WT CG15117 enhancer and after 14 mutations. The CG15117 enhancer showed a slightly altered T1 pattern following incorporation of the KC code. g, Nucleotide contribution scores of WT Pkc53e enhancer sequence and after 9 mutations for T2 (top), T1 (middle) and γ-KC (bottom). In b, e, the expected location of KC is shown with dashed circles. Scale bars, 100 µm. In d,f,g, the position of the mutations is shown with dashed lines, the mutational order is written in between top and bottom plots and motif annotation is indicated with strong (s) or weak (w) motif instances.