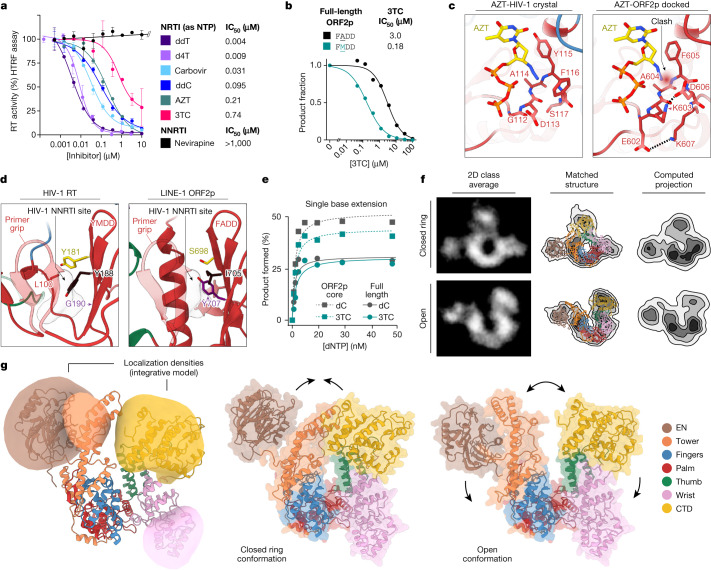
Fig. 4. Inhibition and structure of full-length ORF2p.
a, The ORF2p core was inhibited by NRTIs but not allosteric NNRTI HIV inhibitors in vitro according to homogeneous time-resolved fluorescence assay (n = 3 wells). b, 3TC inhibition in gel-based RT assay of full-length ORF2p WT (FADD) or HIV-like (FMDD). Although both were efficient RTs, 3TC more potently inhibited HIV-like FMDD than WT ORF2p. c, Structural basis for poor L1 inhibition by AZT. Crystal structure of AZT triphosphate bound to HIV-1 RT (PDB 5I42) versus model of AZT triphosphate bound to L1 ORF2p. A clash between the 3′-azido and ORF2p F605 backbone NH is highlighted. Dashed lines indicate salt bridges rigidifying the ORF2p pocket. d, Comparison of the HIV-1 RT NNRTI-binding region with ORF2p. Left, HIV-1 RT in the NNRTI-unbound conformation (PDB 7LRI). Residues involved in NNRTI-resistance are highlighted; space occupied by HIV-1-bound nevirapine is shadowed (PDB 4PUO). Right, equivalent region in L1 ORF2p. The long α-helix corresponds to residues 572–588 in ORF2p. Residues analogous to those in HIV-1 RT are labelled. e, Quantification of single-nucleotide incorporation RT assay showing that purified ORF2p core and full-length ORF2p are similarly active in incorporation of dC or 3TC nucleotides. f,g, Integrative modelling of the full-length ORF2p using Integrative Modeling Platform software, combining data from AlphaFold, molecular dynamics simulations, cryo-EM and cross-linking mass spectrometry generated an ensemble of conformational states. f, Negative stain transmission electron microscopy validation: class averages were postprocessed and matched to projection images of ORF2p models. g, Localization densities represent the structural flexibility of EN, tower, wrist and CTD domains in the ensemble of full-length ORF2p models. Representative full-length ORF2p models from the validated ensemble highlight concerted movements of EN, tower and CTD relative to fingers, palm and thumb, together allowing ORF2p to adopt open and closed states. Data in a, b and e are representative of two independent experiments and shown as mean ± s.d.