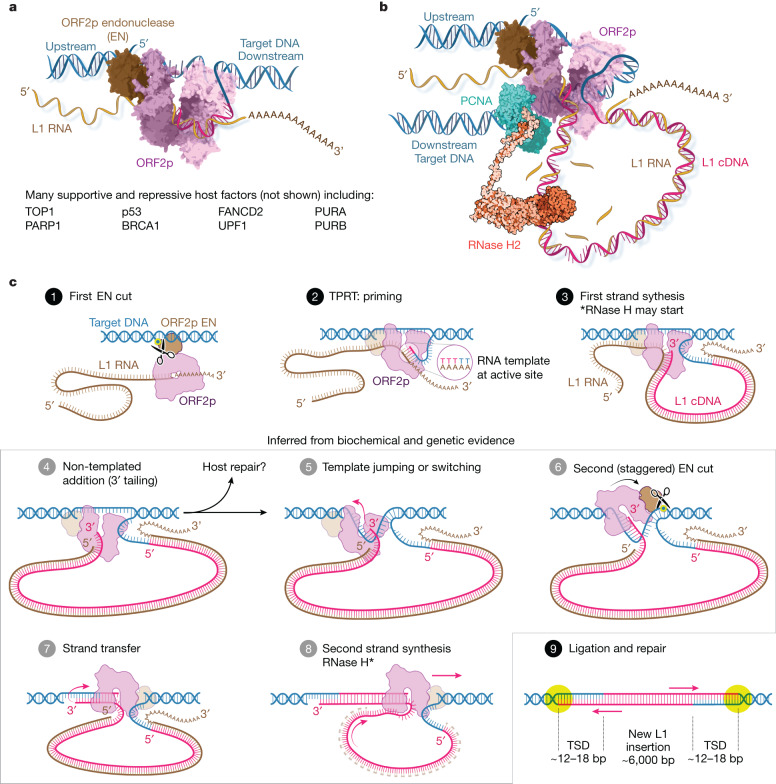
Fig. 6. Revised L1 insertion model.
a, ORF2p bound to target DNA as TPRT begins, drawn schematically with linear target DNA for clarity as in the models below. b, ORF2p in complex during first strand synthesis. It seems more likely that ORF2p bends the target DNA around the highly positively charged ‘back’ face of the polymerase (Extended Data Fig. 9); it can then pass through the PCNA ring clamp, which binds to the PIP box and recruits RNase H2 (ref. 29). c, Revised insertion model. Activities supporting steps 4, 5, 7 and 8 are demonstrated here. 1. ORF2p EN cuts target DNA, liberating a gDNA 3′-OH 2. TPRT: the T-rich gDNA primer is passed into the RT active site, where it base pairs with the poly(A) tail of the bound template, and the 3′-OH is extended. 3. First strand synthesis generates a large (6 kb) cDNA loop; RNase H2, recruited by ORF2p–PCNA, can begin. 4. NTA, in which extra bases are added to the 3′ cDNA end beyond the 5′ end of the RNA template, may occur. 5. Template jumping or switching to the exposed single-stranded gDNA may follow, potentially facilitated by microhomology from NTA nucleotides and the 5′ cap. This would also release 5′ phosphate-bound EN to ‘rock and roll’20,24,48 to carry out: 6. The second EN (staggered) cut, which liberates the 3′ OH used to prime second strand synthesis; a stagger from the first cut of approximately 12–18 bp results in characteristic target site duplications (TSDs)20,21,24,44. 7. Strand transfer and priming of second strand synthesis. 8. Second strand synthesis using the 6 kb L1 cDNA as template. RNase H2 activity may also occur here. 9. Ligation and end repair, resulting in a completed approximately 6 kb insertion flanked by TSDs. The second EN cleavage may sometimes occur in the absence of a template jump. b, © 2023 JHUAAM. Illustration: Jennifer E. Fairman.