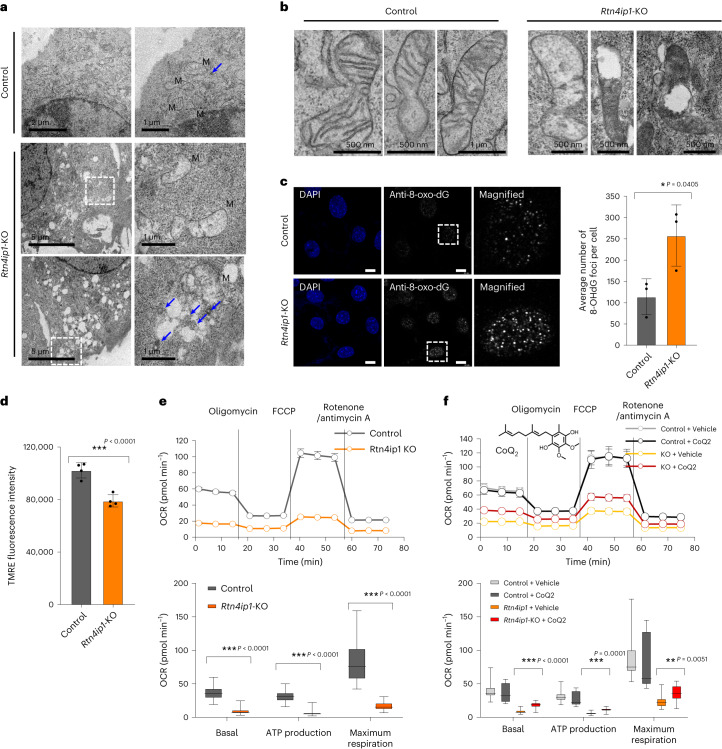
Fig. 5. RTN4IP1/OPA10 regulates oxidative stress and mitochondrial respiration.
a, TEM images of control (upper panel) and Rtn4ip1-knockout (KO) (lower panel) C2C12 cells. Mitochondrial structures are marked with ‘M’ and multilamella body structures are marked with blue arrows. b, Magnified TEM images of control (left panel) and Rtn4ip1-KO (right panel) C2C12 cells focusing on the mitochondria. c, Confocal microscopy images of control and Rtn4ip1-KO C2C12 cells. Scale bars, 10 µm. DNA regions oxidized by intracellular ROS were stained with an anti-8-oxo-dG monoclonal antibody and total DNA was stained with 4,6-diamidino-2-phenylindole (DAPI). The average number of 8-OHdG foci per cell was counted in images from control and Rtn4ip1-KO C2C12 cells (n = 3 biological replicates). d, Mitochondrial membrane potential measurement of control and Rtn4ip1-KO C2C12 cells by TMRE fluorescence. Mean fluorescence of the C2C12 cells in each sample was measured by a microplate reader (n = 4 biological replicates). e, Measurement of OCR, basal respiration, maximal respiration and ATP production in control and Rtn4ip1-KO C2C12 cells (n = 45 biological replicates). f, Rescued OCR of Rtn4ip1-KO cells by CoQ2. Basal respiration, ATP production and maximal respiration of CoQ2-treated Rtn4ip1-KO cells (n = 14 biological replicates). Mean values are shown with error bars representing the standard deviation. Statistical significance was determined using a two-tailed Student’s t-test: *P < 0.05, **P < 0.01, ***P < 0.001. Source data can be found in the Source Data file.