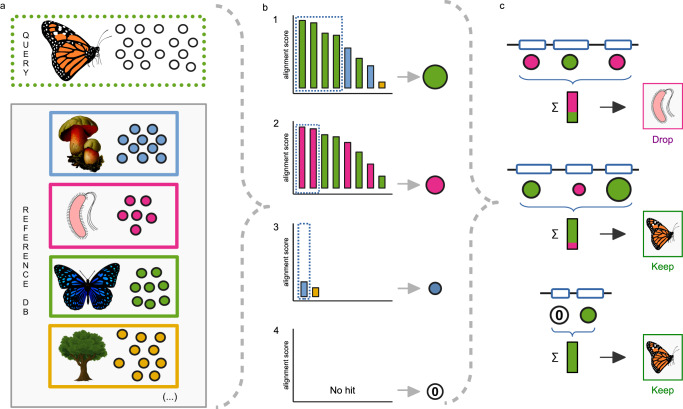
Fig. 1. Overview of the ContScout algorithm.
a A quick database search with the query proteins is performed against a taxonomy-aware reference database. The circles represent individual proteins whose color correspond to different taxonomic lineages: green=metazoa, blue=fungi, purple=bacteria, orange=viridiplantae. The expected lineage one of the query genome (metazoa) is shown as a dotted green frame. Each colored frame with a group of colored dots and a thumbnail image represents one of the many reference genomes in the database. b The bar charts, illustrating cases 1-4, show query versus reference database alignment scores ranked in decreasing order. Taxon information of the best hit is assigned to each query protein together with a confidence score (proportional to dot size). Protein-wise taxon call examples: Case 1: many hits support the metazoa (green) taxon label that is assigned with a high confidence score. Case 2: bacteria (purple) taxon label is assigned but due to limited support, the confidence score is lower. Case 3: Fungi label (blue) from a single hit is assigned albeit with a very low confidence score. Case 4: No hit observed for query in the reference database. c Protein taxon votes are summarized over contigs / scaffolds (∑ sign) and turned into consensus contig calls based on the user-defined threshold. When the consensus taxon label of a contig / scaffold disagrees with that of the query genome, the contig is removed, together with all associated proteins.