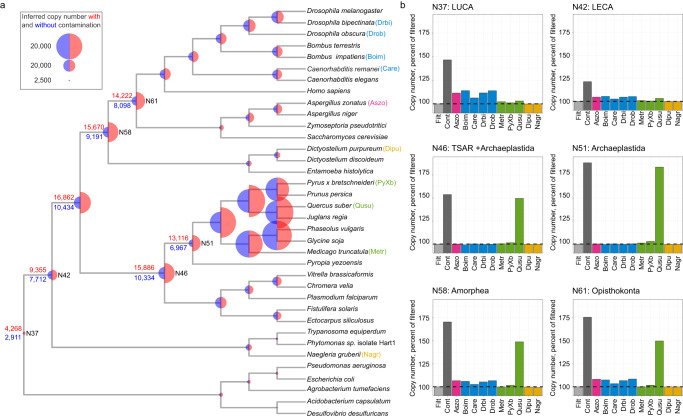
Fig. 6. Effect of contamination on ancestral genome reconstruction in the 36 genome data set.
a Proportionally scaled, color-coded semi-circles correspond to copy number estimates, calculated by Compare22 with the red color representing contaminated and blue color standing for cleaned data. Additionally, the copy numbers for seven prominent internal nodes are provided within the species tree as text node labels. b Bar plots show the bias of individual contaminated genomes introduced to copy number prediction at six selected internal nodes (N37: LUCA, N42: LECA, N46: TSAR+Archaeplastida, N51: Archaeplastida, N58:Amorphea, N61:Opisthokonta), where 100% corresponds to the copy number inferred from clean data. Abbreviations used on bar plot legends: Filt: decontaminated, Cont: contaminated. Species name abbreviation key is provided as part of the species tree leaf node labels in panel a. Bar plot are colored according to taxon lineages of the genomes with blue color representing animals, purple corresponding to fungi, green standing for plants and orange for other eukaryotes. Bars corresponding to clean data are colored in light grey, while those ones that stand for unfiltered data are charted in dark grey.