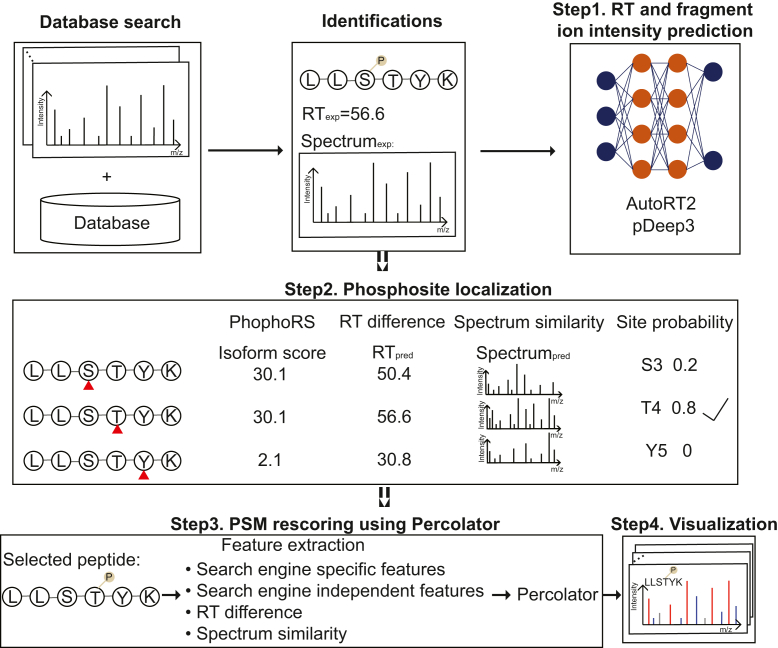
Fig. 1.
DeepRescore2 workflow for phosphopeptide identification and phosphosite localization. After searching the MS/MS spectra against a protein database using a search engine, DeepRescore2 processes the search results in four steps: (1) RT and fragment ion intensity prediction for all identified peptide sequences with all possible phosphosite localizations using deep learning models fine-tuned with confidently identified PSMs from the database search. (2) Phosphosite localization by combining site localization score from PhosphoRS and deep learning prediction derived spectrum similarity and retention time (RT) difference scores. (3) PSM rescoring using Percolator based on search engine specific features, search engine independent features, and spectrum similarity and retention time difference scores. (4) Visualization of the PSMs by PDV.