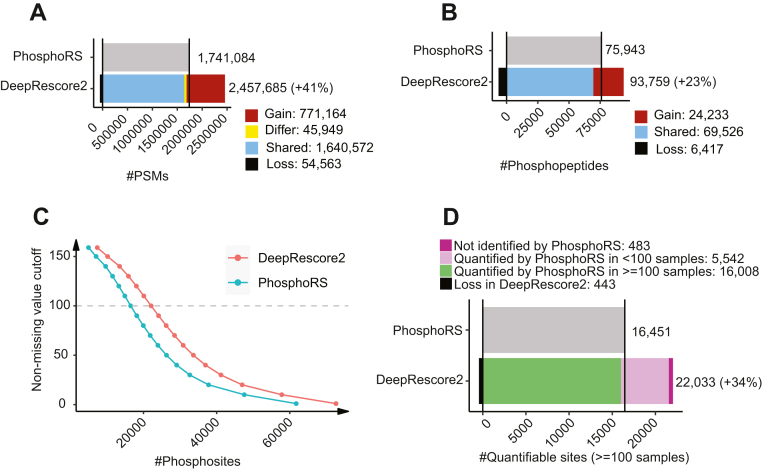
Fig. 5.
Performance evaluation on a large liver cancer dataset.A and B, the numbers PSMs (A) and phosphopeptides (B) identified by MaxQuant in combination with phosphoRS or DeepRescore2, respectively. Gain: identified by DeepRescore2 but not PhosphoRS. Shared: identified by both DeepRescore2 and PhosphoRS. Loss: identified by PhosphoRS but not DeepRescore2. Differ: different sites identified by DeepRescore2 and PhosphoRS, respectively. C, the numbers of “quantifiable” phosphosites based on different levels of non-missing value cutoff, i.e., numbers of samples with a non-missing value in tumors and NATs, respectively. D, the numbers of quantifiable phosphosites (i.e., quantified in at least 100 tumor samples and 100 NAT samples) in the data tables generated by DeepRescore2 and PhosphoRS, respectively.